Back to Journals » International Journal of General Medicine » Volume 14
A Novel Pyroptosis-Associated Long Noncoding RNA Signature to Predict the Prognosis of Patients with Colorectal Cancer
Received 14 July 2021
Accepted for publication 17 September 2021
Published 27 September 2021 Volume 2021:14 Pages 6111—6123
DOI https://doi.org/10.2147/IJGM.S328842
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Sijun Chen, Jianwei Zhu, Xiaofei Zhi
Department of General Surgery, Affiliated Hospital of Nantong University, Nantong, 226001, Jiangsu, People’s Republic of China
Correspondence: Xiaofei Zhi
Department of General Surgery, Affiliated Hospital of Nantong University, Nantong, 226001, Jiangsu, People’s Republic of China
Tel +86 18851323088
Email [email protected]
Purpose: Pyroptosis plays an important role in tumor progression. However, there is no pyroptosis-associated long noncoding RNA (lncRNA) signature to predict the prognosis of patients with colorectal cancer (CRC).
Materials and Methods: The RNA sequencing data (RNA-seq) and corresponding clinical information relating to CRC patients were obtained from the Cancer Genome Atlas (TCGA) database and the GSE39582 dataset. Univariate Cox regression analysis was used to identify pyroptosis-associated lncRNAs linked to CRC prognosis. Subsequently, multivariate Cox regression analysis was performed to construct a pyroptosis-associated lncRNAs signature within the TCGA cohort, which was then validated using the GSE39582 dataset. We used Kaplan–Meier (K-M) analysis, principal component analysis (PCA), and receiver operating characteristic curve (ROC) analysis to evaluate our novel lncRNA signature. Finally, gene set enrichment analysis (GSEA) was performed to explore the potential function of the lncRNA signature.
Results: We constructed a pyroptosis-associated lncRNA signature comprising four lncRNAs (ELFN1-AS1, PCAT6, TNRC6C-AS1, and ZEB1-AS1). CRC patients were subdivided into high- and low-risk groups based on median risk scores. The results of the K-M, PCA, and ROC analyses showed that this signature could accurately predict the prognosis of CRC patients. Univariate and multivariate Cox regression analyses showed that the pyroptosis-associated signature was an independent prognostic factor. Functional analysis suggested that tumor-associated pathways were enriched for in the high-risk CRC patient group.
Conclusion: Our study established an effective prognostic signature for CRC patients that may represent a potential therapeutic target.
Keywords: colorectal cancer, gene signature, overall survival, pyroptosis, lncRNA
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related deaths worldwide, with an incidence that is rising year by year among adolescents.1,2 The early symptoms of CRC are subtle. Patients therefore often present at an advanced stage by the time CRC is diagnosed.3 The 5-year survival rate of CRC patients diagnosed in the early stages is > 90%, while the 5-year survival rate of CRC patients diagnosed with metastatic cancer is only 12%.4,5 Surgery and chemotherapy are currently the main treatment methods for colorectal cancer, and the treatment efficacy for advanced patients remains poor.6 There is still a shortage of simple and effective methods for the prediction of CRC prognosis. It is therefore imperative to explore the underlying developmental mechanisms implicated in CRC and identify effective prognostic biomarkers.
Pyroptosis is a newly-discovered form of programmed cell death that is predominantly induced by certain inflammasomes,7 such as the caspase-1/4/5/11 inflammasome pathways.8 The main manifestations of pyroptosis are cell swelling, cell membrane rupture, cell membrane pore formation, and the release of proinflammatory contents.9,10 Pyroptosis is strongly associated with clinical abnormalities such as cancer, Alzheimer’s disease, systemic lupus erythematosus (SLE), renal ischemia reperfusion injury (IRI), and diabetes.11–14 The induction of cancer cell death via pyroptosis represents a novel method for the treatment of tumors resistant to traditional therapies.15 In recent years, some researchers have studied pyroptosis-associated pathways to find new targeted treatment strategies for CRC.
Long noncoding RNA (lncRNA) is a type of RNA >200 nucleotides in length, which lacks the capacity to encode proteins.16 LncRNAs mediate multiple biological functions, such as transcription, translation, chromatin modifications, and allosteric regulation of enzyme activities.17 Numerous studies have suggested that pyroptosis is regulated by many factors, including lncRNAs.18 However, few studies documenting the existence of pyroptosis-associated lncRNAs and their roles in CRC prognosis exist to date, and this is an area of research that needs further investigation.
In this study, we constructed a pyroptosis-associated lncRNA signature and validated its function in an independent cohort of CRC patients. We demonstrate that this novel signature can predict the prognosis of CRC, which could lead to the identification of therapeutic targets and biomarkers.
Materials and Methods
Data Sources
Figure 1 shows our analysis workflow. The RNA sequencing data (RNA-seq) and corresponding clinical information associated with the training cohort were downloaded from the Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) database. The training cohort contains 473 colon adenocarcinoma samples, one of the most common types of CRC.19 The GSE39582 dataset, which was obtained from the gene expression omnibus (GEO: https://www.ncbi.nlm.nih.gov/geo/) database, was used as a testing cohort. All RNA-seq data were normalized with the log2(x+1) transformation. A total of 133 pyroptosis-associated genes were extracted from the GeneCards (https://www.genecards.org/) database and are outlined in Supplementary Table S1.
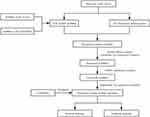 |
Figure 1 Flowchart outlining the research process. The flowchart depicts the process of pyroptosis-associated lncRNA signature identification and validation. |
Identification of Pyroptosis-Associated lncRNAs
LncRNA and mRNA were identified based on the annotations provided by Ensembl Genomes (https://ensemblgenomes.org/). Shared lncRNAs were obtained from the two cohorts and a Venn diagram was generated using the “Venn” R package. Pearson correlation analysis was performed to analyze the correlation between shared lncRNAs and pyroptosis-associated genes in CRC. LncRNAs with correlation coefficients |R2| > 0.3 and P < 0.001 were considered to be pyroptosis-associated lncRNAs.
Construction of a Pyroptosis-Associated lncRNA Signature
Univariate Cox regression and Kaplan–Meier (K-M) analyses were used to identify pyroptosis-associated lncRNAs linked to overall survival (OS), and lncRNAs with P < 0.05 were considered statistically significant for further analysis. In order to avoid over-fitting, the candidate lncRNAs were screened by the least absolute shrinkage and selection operator (LASSO) regression analysis in the “glmnet” R package. Subsequently, these candidate lncRNAs were included in the multivariate Cox regression analysis and the optimal lncRNA signature was constructed based on the lowest Akaike information criterion (AIC) value. The risk score of each patient was calculated according to the following formula: Risk score=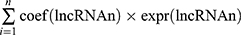 . The patients were subdivided into high-risk groups and low-risk groups based on the median risk score.
. The patients were subdivided into high-risk groups and low-risk groups based on the median risk score.
Validation of the Pyroptosis-Associated lncRNA Signature
K-M survival analysis was performed to compare the OS between the high and low-risk CRC patient groups. Receiver operating characteristic (ROC) and Cox regression analyses were further used to analyze the prognostic value of our lncRNA signature. Principal component analysis (PCA) was used to explore the distribution of the different groups. The testing cohort was used to validate the signature. The “rms” R package was used to establish a nomogram.
Enrichment Analysis
Gene set enrichment analysis (GSEA) is a calculation method used to determine the functional difference between two groups. GSEA software (version 4.1.0) was used to analyze the enrichment of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway in high and low-risk CRC groups.
Statistical Analysis
All statistical analyses were conducted using R software (version 4.0.3). The lncRNAs and mRNAs co-expression network was drawn using CYTOSCAPE software (version 3.7.1). A t-test was used for the clinical correlation analysis. Two-tailed P < 0.05 was considered as a measure of statistical significance.
Results
Identification of Pyroptosis-Associated lncRNAs
A total of 14,142 lncRNAs were identified from the TCGA dataset and 1146 lncRNAs were screened from the GSE39582 dataset. After deleting the duplicate lncRNAs in each database, we obtained 1139 shared lncRNAs (Figure 2A). We formed a co-expression network of shared lncRNAs and pyroptosis-associated genes in the training cohort. Among the shared lncRNAs, 174 lncRNAs were considered to be associated with pyroptosis, determined using Pearson correlation analysis. (|R2| > 0.3 and P < 0.001).
Construction of a Pyroptosis-Associated lncRNA Signature in CRC
Univariate Cox regression and K-M analyses were performed to screen prognostic lncRNAs within the training cohort (Figure 2B). In order to avoid over-fitting, the lncRNAs with P < 0.05 were included in the LASSO regression analysis to obtain five candidate lncRNAs (Figure 2C and D). Next, multivariate Cox regression analysis was applied to the candidate lncRNAs (Figure 2E). Based on the optimal AIC value (AIC=905.46), we constructed a lncRNA signature comprising four pyroptosis-associated lncRNAs (ELFN1-AS1, PCAT6, TNRC6C-AS1, and ZEB1-AS1). These four lncRNAs were associated with an unfavorable prognosis in CRC patients and were considered risk factors (HR>1). We built a co-expression network to show the relationship between pyroptosis-associated lncRNAs and mRNAs (Figure 3).
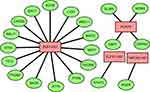 |
Figure 3 Co-expression network of pyroptosis genes and lncRNAs. Green nodes indicate pyroptosis genes and red nodes represent lncRNAs. |
Validation of the Pyroptosis-Associated lncRNA Signature
The median risk score was used to dichotomize patients into a high- (n=223) and a low-risk (n=223) groups in the training cohort. K-M survival analysis indicated the OS of CRC patients in the high-risk group was worse than the low-risk group (P < 0.05; Figure 4A). Time-dependent ROC curves demonstrated that the signature has a reliable prognostic value. The area under the ROC curve (AUC) at 1-, 3-, and 5-years reached 0.700, 0.698, and 0.674, respectively (Figure 4B). PCA illustrated that CRC patients in the high- and low-risk groups were distributed in different directions (Figure 4C). The risk curve and scatterplot were applied to illustrate the risk score in the low- and high-risk groups, and the relevant survival status of CRC patients (Figure 4D and E, low-risk group to the left side of the dotted line and the high-risk group to the right side of the dotted line). The results suggested that the mortality rates were dependent on the risk score. The heatmap showed that these four lncRNAs were upregulated in the high-risk group (Figure 4F).
To further evaluate the accuracy of the lncRNA signature in predicting CRC prognosis, we used the same methods to validate it in the testing cohort extracted from the GSE39582 dataset. K-M analysis shows that patients in the high-risk group also had a worse prognosis than those in the low-risk group (P < 0.05, Figure 5A). Meanwhile, the AUCs for predicting 1-, 3-, and 5-year survival rates were 0.648, 0.608, and 0.601, respectively (Figure 5B). In addition, CRC patients from the testing cohort were also distributed in two directions in the PCA (Figure 5C). The distribution of the risk score and survival status in the training cohort are presented in Figure 5D and E. More deaths and shorter survival times were recorded for the high-risk group, compared to the low-risk group. The expression differences of the four lncRNAs in the high- and low-risk groups were also similar to those in the training cohort (Figure 5F). In general, these results offer evidence of the robustness and effectiveness of our signature.
The Clinical Value of the Pyroptosis-Associated lncRNA Signature
Univariate and multivariate Cox regression analyses were performed to assessed whether the risk score was an independent prognostic factor in the context of CRC. The univariate Cox regression analysis indicated that the risk score was a factor predicting survival in both the training and testing cohorts (P < 0.05, Figure 6A and C). The multivariate Cox regression analysis also implied that, after eliminating the influence of clinical features (ie gender, age, clinical stage, and TNM stage), the risk score was an independent prognostic factor in both cohorts. (P < 0.05, Figure 6B and D).
 |
Figure 6 Univariate and multivariate Cox regression analyses of the risk scores and clinical characteristics relating to OS in the training cohort (A and B) and the testing cohort (C and D). |
To further assess the clinical value of our lncRNA signature, we analyzed the correlation between risk score and other clinical factors using a t-test. As shown in Table 1, the risk score was closely related to clinical characteristics, such as stage, tumor metastasis, and lymphatic metastasis, in the training cohort (P < 0.05). The risk score was also found to be significantly related to stage, tumor size, tumor metastasis, and lymphatic metastasis in the testing cohort (Table 2). These results suggest that the four pyroptosis-associated lncRNAs participate in the development of CRC.
 |
Table 1 Relationship Between Clinical Features and the Risk Score in the Training Cohort |
 |
Table 2 Relationship Between Clinical Features and the Risk Score in the Testing Cohort |
Establishment of the Nomograph
Based on these clinical characteristics, we constructed a nomograph in the TCGA cohort to help professionals make appropriate clinical decisions when treating CRC patients (Figure 7A). The nomograph showed that the risk score is the main factor affecting the prognosis of CRC patients. The projected values of the 1-, 3-, and 5-year calibration curves were very close to the actual values (Figure 7B–D), showing that the nomograph we established was accurate and dependable.
Gene Set Enrichment Analysis
The GSEA was performed to explore further functional differences between high- and low-risk groups in the TCGA cohort. The gene sets with a screening criterion of P < 0.05 were considered to be significantly enriched. The high-risk group was mainly enriched for activated tumor-associated pathways (Figure 8A). Surprisingly, the multiple metabolic pathways enriched in the low-risk group were associated with pyroptosis (Figure 8B). Of note, the glucose metabolic and leucine metabolic pathways have been reported to be closely linked to pyroptosis.20,21
Discussion
CRC is a common type of malignant tumor. At present, the treatment options for CRC include surgery, chemotherapy, and radiotherapy. However, these methods are far from satisfactory, and to date, there is no effective method to accurately predict the prognosis of CRC patients. The aim of the present study was to identify a new and effective diagnostic signature, which could identify high-risk CRC patients at an earlier stage enabling initiation of appropriate treatment, thus potentially changing the course and outcome of the disease.
Pyroptosis, a word derived from the Greek roots “pyro” and “ptosis”, which literally translated to “fever” and “falling”, is the name given to this newly discovered type of inflammatory programmed cell death.22 Pyroptosis is rapidly emerging as an area of focus in cancer research, and recent studies have found that pyroptosis plays an important role in tumorigenesis.23 Therefore, the biomarkers associated with pyroptosis are expected to yield new therapeutic targets and diagnostic biomarkers. Recent studies have found that lncRNA is one of the factors that regulate pyroptosis.24,25 Here, we firstly establish a pyroptosis-associated lncRNA signature in the context of CRC, before evaluating its prognostic value.
In this study, we identified shared lncRNAs from two databases to ensure that the lncRNAs used to construct the signature were universal. Pyroptosis-associated lncRNAs were initially identified by constructing a co-expression network of pyroptosis genes and lncRNAs. Further, multivariate Cox regression analyses was performed to construct a pyroptosis-associated lncRNA signature in the training cohort. CRC patients were subdivided into high- and low-risk groups based on their risk scores, with the CRC patients in the high-risk group exhibiting a worse prognosis. The found that our lncRNA signature was capable of predicting the prognosis of CRC patients with a high level of accuracy in but the training and testing cohorts. In addition, we identified the risk score as an independent prognostic factor in relation to tumor progression. To help clinical decision making, we constructed a nomogram, which confirmed that the lncRNA signature was an independent predictor of CRC survival.
To further explore the biological function of lncRNAs in CRC, we consulted the existing literature. We found that the presence of the four pyroptosis-associated lncRNAs correlated with tumor progression. For instance, a previous study has shown that ELFN1-AS1 promotes CRC by up-regulating TRIM44 by acting as a sponge for miR-4644,26 which is consistent with our results. Furthermore, TNRC6C-AS1 was found to promote the proliferation, apoptosis, migration, and invasion of thyroid carcinoma by suppressing the expression of TNRC6C.27 However, there is no prior evidence relating to its involvement in CRC. Our study has for the first time shown that TNRC6C-AS1, which is a component of the novel lncRNA signature, acts as an independent predictor of CRC.
Previous work has shown that ZEB1-AS1 promotes CRC cell proliferation by regulating Wnt/β-catenin signaling,28 while PCAT6 is one of the most commonly reported lncRNA oncogene regulatory factors and its expression is upregulated in a variety of tumors, such as cholangiocarcinoma, glioblastoma, ovarian cancer, and colorectal cancer.29–32 Huang et al32 found that PCAT6 forms a complex with EZH2 to inhibit cell apoptosis and promote the progression of CRC. In addition, PCAT6 regulates 5-fluorouracil resistance through HMGA2 signaling by targeting miR-204 in CRC cells.33 The above research implies that these lncRNAs can be used as prognostic indicators and may be used as potential therapeutic targets in cancer. However, how these lncRNAs regulate cancer cells via pyrolysis still needs further study. Our GSEA results showed that the Notch signaling pathway, which is implicated in the development progress of cancer,34 was enriched in the high-risk CRC group. The Notch signaling pathway plays a critical role in cancer invasion and metastasis through crosstalk with other signaling pathways.35
Despite our important findings, we acknowledge that there are some limitations in our research. Firstly, more experiments are needed to verify the relationship between the expression of lncRNAs and CRC cells. Secondly, the mechanism employed by pyroptosis-associated lncRNAs need to be further explored.
Conclusion
Our study describes the construction of a novel pyroptosis-associated lncRNA signature, which can be used as a biomarker to accurately predict the prognosis of CRC patients. More importantly, the signature may provide new insights into the treatment of CRC patients.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81702369), and the China Postdoctoral Science Foundation Grant (2016M601868 and 2018T110534).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Mariotto AB, Yabroff KR, Shao Y, Feuer EJ, Brown ML. Projections of the cost of cancer care in the United States: 2010–2020. J Natl Cancer Inst. 2011;103(2):117–128. doi:10.1093/jnci/djq495
2. Weinberg BA, Marshall JL, Salem ME. The growing challenge of young adults with colorectal cancer. Oncology. 2017;31(5):381–389.
3. Siegel RL, Jakubowski CD, Fedewa SA, Davis A, Azad NS. Colorectal cancer in the young: epidemiology, prevention, management. Am Soc Clin Oncol Educ Book. 2020;40:1–14. doi:10.1200/EDBK_279901
4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi:10.3322/caac.21254
5. Chen L, Lu D, Sun K, et al. Identification of biomarkers associated with diagnosis and prognosis of colorectal cancer patients based on integrated bioinformatics analysis. Gene. 2019;692:119–125. doi:10.1016/j.gene.2019.01.001
6. Bertelsen CA, Bols B, Ingeholm P, Jansen JE, Neuenschwander AU, Vilandt J. Can the quality of colonic surgery be improved by standardization of surgical technique with complete mesocolic excision?. Colorectal Dis. 2011;13(10):1123–1129. doi:10.1111/j.1463-1318.2010.02474.x
7. Ruan J, Wang S, Wang J. Mechanism and regulation of pyroptosis-mediated in cancer cell death. Chem Biol Interact. 2020;323:109052. doi:10.1016/j.cbi.2020.109052
8. Wang YY, Liu XL, Zhao R. Induction of pyroptosis and its implications in cancer management. Front Oncol. 2019;9:971. doi:10.3389/fonc.2019.00971
9. Fink SL, Cookson BT. Pyroptosis and host cell death responses during Salmonella infection. Cell Microbiol. 2007;9(11):2562–2570. doi:10.1111/j.1462-5822.2007.01036.x
10. Fink SL, Bergsbaken T, Cookson BT. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc Natl Acad Sci USA. 2008;105(11):4312–4317. doi:10.1073/pnas.0707370105
11. Tan MS, Tan L, Jiang T, et al. Amyloid-beta induces NLRP1-dependent neuronal pyroptosis in models of Alzheimer’s disease. Cell Death Dis Aug. 2014;5(8):e1382. doi:10.1038/cddis.2014.348
12. Magna M, Pisetsky DS. The role of cell death in the pathogenesis of SLE: Is pyroptosis the missing link? Scand J Immunol. 2015;82(3):218–224. doi:10.1111/sji.12335
13. Wu H, Huang T, Ying L, et al. MiR-155 is involved in renal ischemia-reperfusion injury via direct targeting of Foxo3a and regulating renal tubular cell pyroptosis. Cell Physiol Biochem. 2016;40(6):1692–1705. doi:10.1159/000453218
14. Lin CF, Kuo YT, Chen TY, Chien CT. Quercetin-rich guava (Psidium guajava) juice in combination with trehalose reduces autophagy, apoptosis and pyroptosis formation in the kidney and pancreas of type II diabetic rats. Molecules. 2016;21(3):334. doi:10.3390/molecules21030334
15. Yu J, Li S, Qi J, et al. Cleavage of GSDME by caspase-3 determines lobaplatin-induced pyroptosis in colon cancer cells. Cell Death Dis. 2019;10(3):193. doi:10.1038/s41419-019-1441-4
16. Fazal FM, Chang HY. lncRNA structure: message to the heart. Mol Cell. 2016;64(1):1–2. doi:10.1016/j.molcel.2016.09.030
17. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62. doi:10.1038/nrg.2015.10
18. Chen Z, He M, Chen J, Li C, Zhang Q. Long non-coding RNA SNHG7 inhibits NLRP3-dependent pyroptosis by targeting the miR-34a/SIRT1 axis in liver cancer. Oncol Lett. 2020;20(1):893–901. doi:10.3892/ol.2020.11635
19. Mohamed A, Menon H, Chulkina M, Yee NS, Pinchuk IV. Drug-microbiota interaction in colon cancer therapy: impact of antibiotics. Biomedicines. 2021;9(3):259. doi:10.3390/biomedicines9030259
20. Jin H, Zhu Y, Wang XD, et al. BDNF corrects NLRP3 inflammasome-induced pyroptosis and glucose metabolism reprogramming through KLF2/HK1 pathway in vascular endothelial cells. Cell Signal. 2021;78:109843. doi:10.1016/j.cellsig.2020.109843
21. Fenini G, Karakaya T, Hennig P, Di Filippo M, Beer HD. The NLRP1 inflammasome in human skin and beyond. Int J Mol Sci. 2020;21(13):4788. doi:10.3390/ijms21134788
22. Cookson BT, Brennan MA. Pro-inflammatory programmed cell death. Trends Microbiol. 2001;9(3):113–114. doi:10.1016/s0966-842x(00)01936-3
23. Ye J, Zhang R, Wu F, et al. Non-apoptotic cell death in malignant tumor cells and natural compounds. Cancer Lett. 2018;420:210–227. doi:10.1016/j.canlet.2018.01.061
24. Yang F, Qin Y, Wang Y, et al. LncRNA KCNQ1OT1 mediates pyroptosis in diabetic cardiomyopathy. Cell Physiol Biochem. 2018;50(4):1230–1244. doi:10.1159/000494576
25. Wan P, Su W, Zhang Y, et al. LncRNA H19 initiates microglial pyroptosis and neuronal death in retinal ischemia/reperfusion injury. Cell Death Differ. 2020;27(1):176–191. doi:10.1038/s41418-019-0351-4
26. Lei R, Feng L, Hong D. ELFN1-AS1 accelerates the proliferation and migration of colorectal cancer via regulation of miR-4644/TRIM44 axis. Cancer Biomark. 2020;27(4):433–443. doi:10.3233/CBM-190559
27. Muhanhali D, Zhai T, Jiang J, Ai Z, Zhu W, Ling Y. Long non-coding antisense RNA TNRC6C-AS1 is activated in papillary thyroid cancer and promotes cancer progression by suppressing TNRC6C expression. Front Endocrinol. 2018;9:360. doi:10.3389/fendo.2018.00360
28. Lv SY, Shan TD, Pan XT, et al. The lncRNA ZEB1-AS1 sponges miR-181a-5p to promote colorectal cancer cell proliferation by regulating Wnt/beta-catenin signaling. Cell Cycle. 2018;17(10):1245–1254. doi:10.1080/15384101.2018.1471317
29. Xin Y, He X, Zhao W, et al. LncRNA PCAT6 increased cholangiocarcinoma cell proliferation and invasion via modulating miR-330-5p. Am J Transl Res. 2019;11(9):6185–6195.
30. Liu P, Zhao P, Li B, Xu D, Wang K. LncRNA PCAT6 regulated by YY1 accelerates the progression of glioblastoma via miR-513/IGF2BP1. Neurochem Res. 2020;45(12):2894–2902. doi:10.1007/s11064-020-03138-4
31. Kong FR, Lv YH, Yao HM, Zhang HY, Zhou Y, Liu SE. LncRNA PCAT6 promotes occurrence and development of ovarian cancer by inhibiting PTEN. Eur Rev Med Pharmacol Sci. 2019;23(19):8230–8238. doi:10.26355/eurrev_201910_19132
32. Huang W, Su G, Huang X, et al. Long noncoding RNA PCAT6 inhibits colon cancer cell apoptosis by regulating anti-apoptotic protein ARC expression via EZH2. Cell Cycle. 2019;18(1):69–83. doi:10.1080/15384101.2018.1558872
33. Wu H, Zou Q, He H, et al. Long non-coding RNA PCAT6 targets miR-204 to modulate the chemoresistance of colorectal cancer cells to 5-fluorouracil-based treatment through HMGA2 signaling. Cancer Med. 2019;8(5):2484–2495. doi:10.1002/cam4.1809
34. Braune EB, Lendahl U. Notch – a goldilocks signaling pathway in disease and cancer therapy. Discov Med. 2016;21(115):189–196.
35. Li L, Tang P, Li S, et al. Notch signaling pathway networks in cancer metastasis: a new target for cancer therapy. Med Oncol. 2017;34(10):180. doi:10.1007/s12032-017-1039-6
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.





