Back to Journals » OncoTargets and Therapy » Volume 13
A Novel lncRNA NONHSAT053785 Acts as an Independent Risk Factor for Intrahepatic Metastasis of Hepatocellular Carcinoma
Authors Li Y , Li G, Chen X, Huang H, Liao L, Yuan T, Deng S
Received 18 March 2020
Accepted for publication 12 May 2020
Published 11 June 2020 Volume 2020:13 Pages 5455—5466
DOI https://doi.org/10.2147/OTT.S254455
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Leo Jen-Liang Su
Yuwei Li,1,* Guangyao Li,2,* Xia Chen,1 Hengliu Huang,1 Ling Liao,1 Tao Yuan,2 Shaoli Deng1
1Department of Laboratory Medicine, Daping Hospital, Army Medical University (Third Military Medical University), Chongqing 400042, People’s Republic of China; 2Department of Hepatobiliary Surgery, Daping Hospital, Army Medical University (Third Military Medical University), Chongqing 400042, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shaoli Deng Email [email protected]
Purpose: Long noncoding RNAs (lncRNAs) in body fluids have been considered as promising novel biomarkers for tumor-related diseases. The present study aimed to investigate the expression level of lncRNA NONHSAT053785 in serum and its correlation with clinical characteristics of hepatocellular carcinoma (HCC) patients.
Methods: The droplet digital PCR (ddPCR) was used to measure the serum levels of NONHSAT053785 in 112 HCC patients, 96 chronic hepatitis B (CHB) patients, and 99 healthy controls (HC). The correlation between NONHSAT053785 and clinical characteristics was analyzed by chi-square test and Spearman correlation test. The risk factors of intrahepatic metastasis (IM) were detected by univariate and multivariate analyses. Furthermore, the diagnostic value of NONHSAT053785 in HCC and its predictive ability in IM were evaluated by the receiver operating characteristic (ROC) curves.
Results: The level of NONHSAT053785 was significantly increased in the serum of HCC patients and was higher in HCC patients with IM as compared to those without. Additionally, the expression level of NONHSAT053785 was significantly related to IM, Child–Pugh classification, and peripheral blood indicators such as liver metabolic enzymes and positively correlated to IM, Barcelona Clinic Liver Cancer (BCLC) staging, and some peripheral blood indicators. Furthermore, the serum NONHSAT053785 was indicated as an independent predictor for IM in the elderly, non-smoking, drinking, and tumor size ≥ 5 cm subjects. The area under the ROC curve (AUC) was 0.801 (P < 0.0001) for diagnosis of HCC and 0.678 (P =0.0015) for predicting IM.
Conclusion: The increase in serum NONHSAT053785 levels was related to an increased risk of IM, and hence, may serve as a novel biomarker for the diagnosis of HCC and the prediction of IM.
Keywords: long noncoding RNA, hepatocellular carcinoma, intrahepatic metastasis, risk factor
Introduction
Hepatocellular carcinoma (HCC) is a highly prevalent tumor and the fourth most common cause of cancer-related deaths worldwide.1 Multiplicity is a major clinical feature of HCC that can spread to other regions of the liver via portal vein invasion, which is referred to as intrahepatic metastasis (IM).2 Several therapeutic methods exist for treating HCC, such as surgical resection, radio-frequency ablation (RFA), transarterial chemoembolization (TACE), systemic chemotherapy, and liver transplant. However, the prognosis of HCC remains unsatisfactory in the long-term. The frequent recurrence of the disease after curative resection is a challenge of HCC treatment. IM, originating from a primary liver tumor, is considered to be one of the origins of recurrent or multifocal liver tumors.3 Therefore, IM indicates a poor prognosis for HCC. However, the molecular characteristics of IM in HCC are yet to be elucidated.
Long noncoding RNAs (lncRNAs) are defined as transcripts >200 nucleotides in length that have not been translated into proteins.4 These have been proposed to carry out diverse functions and represent a large reservoir of potential tumor targets.4,5 The upregulation of urothelial carcinoma associated 1 (UCA1) has been previously documented in several types of tumors, including HCC, bladder cancer, ovarian cancer, melanoma, esophageal cancer, tongue cancer, and lung cancer. The expression level of UCA1 was closely correlated to the progression, tumor size, invasion depth, and stage of cancer.6 Accumulating evidence has demonstrated the regulatory roles of different classes of lncRNAs in HCC related to several etiologies.7,8 Divergent groups of lncRNAs have been implicated in liver carcinogenesis through interactions with DNA, RNA, or proteins.7 LncRNAs have a typical tissue-specific expression pattern and are readily detectable in body fluids due to their high stability. Compared to the other protein biomarkers expressed in various tissues, the lncRNAs are ideal biomarkers.9 Moreover, the serum lncRNAs, such as LINC00161,10 PVT1,11 lncRNA-D16366,12 and RP11-466I1.1, function as biomarkers in HCC.13 However, none of them has a satisfactory diagnostic performance, which could be attributed to a small sample size. Nonetheless, there is a dearth of studies on circulating lncRNAs for predicting IM of HCC.
In a previous study, we used microarray analysis in HCC tissues and paired peritumoral liver tissues and identified 719 differentially expressed lncRNAs that might be involved in the pathogenesis of HCC. A novel lncRNA NONHSAT053785 was one of these differentially expressed lncRNAs with higher fold-change than the remaining; the expression of this lncRNA was verified by quantitative real-time PCR (qRT-PCR) in human tissues.14 In addition, the length of lncRNA NONHSAT053785 (synonym lnc-G6PC-1:1) sequence is 289 nucleotides, and it is a part of the human glucose-6-phosphatase catalytic subunit on Chr17 (NCBI Sequence ID: NG011808.1).15 RNA-Seq data from the Human Body Map shows NONHSAT053785 is primarily expressed in the liver but also in the kidney.16 In the present study, the expression levels of NONHSAT053785 in the serum of HCC patients were detected, and the correlation with the clinical features was analyzed. Also, the diagnostic efficacy and risk factors related to IM were also investigated.
Materials and Methods
Patient Information
A total of 307 participants, including 112 HCC patients, 96 patients with chronic hepatitis B (CHB), and 99 healthy controls (HC), were recruited in this study between April 2018 and December 2019 at the Daping Hospital of the Army Medical University, China. None of the HCC patients received radiotherapy, chemotherapy, or biotherapy before surgery. The diagnosis of HCC was determined by pathological results, and the hepatitis B virus (HBV) infection was diagnosed by clinical laboratory tests. Healthy controls referred to individuals without liver disease or any type of tumor. Written informed consent was obtained from all participants. This study was approved by the Medical Institutional Ethics Committee of the Daping Hospital, and the experiments were performed in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Table 1 lists the clinical characteristics of the participants.
 |
Table 1 Clinical Characteristics of the Study Subjects |
Collection of Serum Samples and Clinical Data
All blood specimens were stored at 4 °C and disposed within 2 h. Serum was collected by centrifugation (3000 ×g, 15 min, 4 °C). The blood specimens of HCC patients were obtained before surgery, and the collected serum was maintained in TRIzol® LS reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocols, at −80 °C prior to total RNA extraction, while those of CHB patients and healthy individuals were subjected to reverse transcription into cDNA (Roche, Germany).
The baseline data of all subjects with respect to age, gender, smoking status, alcohol consumption, HBV infection status, HBV-DNA level, alpha-fetoprotein (AFP) level, carcinoembryonic antigen (CEA) level, ferritin level, liver function test results, blood routine analysis results, and HCC characteristics such as tumor size, histological differentiation, intrahepatic/extrahepatic metastasis, microvascular invasion (MVI), and gross vascular invasion (GVI) were obtained from the medical records. The MVI was determined based on the pathological findings. Preoperative radiological findings of tumor embolus in the portal veins were defined as GVI. The HCC clinical stage was determined based on the Barcelona Clinic Liver Cancer (BCLC) and tumor node metastasis (TNM) classification system.
RNA Extraction and Reverse Transcription
Total RNA was extracted from serum specimens using TRIzol® LS reagent, following the manufacturer’s instructions, and solubilized in 30 μL of RNase-free water. Then, NanoDrop ND-1000 spectrophotometer (Thermo, Wilmington, DE, USA) was used to detect the quantity and purity of the extracted RNA. The RNA sample with an optical density ratio (260/280) of 1.8–2.0 was included in the subsequent study. Total RNA was reverse transcribed using the EvoScript Universal cDNA Master RT reagent Kit (Roche, Germany). The cDNA products were stored at −80 °C until further analysis.
Droplet Digital Polymerase Chain Reaction (ddPCR)
The expression levels of lncRNA were quantified by ddPCR. The 20-µL PCR reaction consisted of 1µL forward primer, 1µL reverse primer, 2 µL probe, 6 µL template/cDNA, and 10 µL ddPCR Supermix (Bio-Rad, Hercules, CA, USA). Droplets were generated using a QX100 droplet generator (Bio-Rad). A 70 µL of ddPCR droplet generation oil was added to the bottom wells of the droplet generation cartridge. The middle wells were loaded with the PCR reaction mixture. A rubber gasket was placed over the cartridge and loaded into the droplet generator. Then, 40 µL emulsion was loaded into each well of a 96-well plate that was subsequently heat-sealed with a foil and the emulsion was cycled to end point as per the manufacturer’s protocol. The amplicons were analyzed using a Bio-Rad QX100 reader (Bio-Rad). The primers used in this study were designed using Primer 5.0 software (Premier, Canada) and synthesized by Sangon Biotech (Shanghai, China). The primers and probe for lncRNA NONHSAT053785 were as follows:
Forward primer, 5ʹ- AGTTATAGATTTACGTCCACTTTAGA–3ʹ;
Reverse primer, 5ʹ- CACCTGAAAACTGATACACTA–3ʹ;
Probe, 5ʹ-CATCGGTCACTTAAACTTGCCTCACA–3ʹ.
Statistical Analysis
All data were analyzed by SPSS 22.0 (Chicago, IL, USA) and GraphPad Prism 8 software (San Diego, CA, USA). The normality of distribution was evaluated using the Shapiro–Wilk and Kolmogorov–Smirnov tests. Mann–Whitney U-test was used to compare the two groups. For comparison among more than two groups, Kruskal–Wallis H or one-way ANOVA test was used. Chi-square test was used to compare the variables represented as frequencies. Spearman correlation coefficient was analyzed to calculate the correlation between the serum levels of NONHSAT053785 and other variables. Univariate and multivariate logistic regression analyses were used to identify the risk factors associated with intrahepatic metastasis. Also, odds ratio (OR) and 95% confidence interval (CI) were calculated. The sensitivity and specificity of NONHSAT053785 and AFP were assessed by the receiver operating characteristic (ROC) curve and the area under ROC curve (AUC). All tests were two-sided, and P<0.05 was considered statistically significant.
Results
Serum Level of NONHSAT053785 Was Increased in HCC Patients
The expression levels of NONHSAT053785 in 112 HCC serum samples collected before surgery, 96 CHB patients’ serum, and 99 healthy controls serum were detected by ddPCR. Results showed that compared to the CHB and HC groups, the serum levels of NONHSAT053785 were significantly increased in HCC patients; however, no significant difference was observed between the CHB and HC groups (P<0.0001, Figure 1).
Correlation Between Serum NONHSAT053785 and Clinical Characteristics of HCC
The correlations between serum levels of NONHSAT053785 and clinical characteristics in HCC patients are listed in Table 2. To analyze the expression of lncRNA NONHSAT053785, the level at the maximum Youden index was used as the cutoff. In this study, significant differences in the serum levels of NONHSAT053785 were observed between subgroups of patients divided according to IM (with or without, P=0.011), Child–Pugh classification (A or B/C, P=0.037), CEA (≤5 or >5 ng/mL, P=0.037), total bilirubin (T-BIL) (≤23 or >23 µmol/L, P=0.006), aspartate transaminase (AST) (≤40 or >40 U/L, P=0.001), alkaline phosphatase (ALP) (≤125 or >125U/L, P=0.001), and monocyte (≤0.6 or >0.6×109/L, P=0.015).
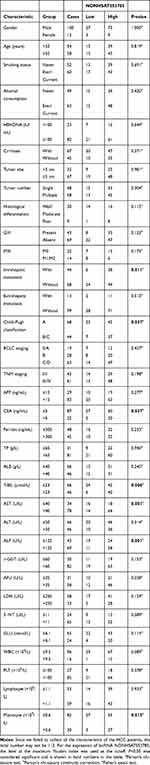 |
Table 2 Correlation Between Serum NONHSAT053785 and Clinical Characteristics of Patients with HCC |
Furthermore, the correlation between intrahepatic metastasis, Child–Pugh classification, and serum levels of NONHSAT053785 in HCC was analyzed by Mann–Whitney U-test. Compared to the patients without IM, NONHSAT053785 expression was significantly increased in patients with IM (P=0.002, Figure 2A). This result suggested that NONHSAT053785 may play crucial roles in HCC invasion ability. However, according to the Child–Pugh classification, no significant difference was observed between HCC patients in stage A and stage B/C (P>0.05, Figure 2B).
Correlation Between Serum NONHSAT053785 and Clinical Characteristics of HCC
We performed Spearman’s analysis to further explore the correlation between serum NONHSAT053785 and the clinical characteristics of HCC (Table 3). In this study, the expression level of serum NONHSAT053785 was positively correlated with IM (r=0.301, P=0.001), BCLC staging (r=0.223, P=0.019), CEA (r=0.205, P=0.033), ferritin (r=0.219, P=0.035), T-BIL (r=0.205, P=0.030), AST (r=0.273, P=0.004), ALP (r=0.362, P<0.0001), γ-glutamyl transferase (γ-GGT) (r=0.260, P=0.006), lactate dehydrogenase (LDH) (r=0.355, P<0.001), 5ʹ-nucleotidase (5ʹ-NT) (r=0.328, P=0.002), white blood cells (WBC) (r=0.378, P<0.0001), and monocytes (r=0.226, P=0.017). The correlation between serum NONHSAT053785 and peripheral blood indexes are shown in Figure 3A–J. These results indicated that NONHSAT053785 may be related to the invasion ability of HCC and abnormal liver metabolism.
 |
Table 3 Spearman’s Analysis of Correlation Between Serum NONHSAT053785 and Clinical Characteristics |
Serum Level of NONHSAT053785 Is an Independent Risk Factor for IM
The relevant risk factors for IM were also identified (Table 4). Univariate analysis suggested that male gender, smoking, alcohol, tumor diameter ≥5 cm, GVI, advanced BCLC stages, advanced TNM stages, and elevated NONHSAT053785 expression were risk factors for IM. These variables were incorporated into a multivariate logistic model to determine whether the expression of serum NONHSAT053785 was an independent risk factor for IM. The TNM staging was not included in multivariate analysis, as it overlapped with BCLC staging. The results showed that tumor diameter ≥5 cm and upregulated NONHSAT053785 expression were independent risk factors of IM.
 |
Table 4 Univariate and Multivariate Analysis for the Risk Factors of Intrahepatic Metastasis |
Stratification Analysis of the Serum Levels of NONHSAT053785 with the Risk of IM
Stratified analysis was performed according to age, smoking status, alcohol consumption, cirrhosis, tumor size, and GVI. As shown in Table 5, we found that the predictive effect of serum NONHSAT053785 on the risk of IM was prominent in elderly (OR= 1.869, 95% CI: 1.049–3.329, P=0.034), non-smoking (OR=1.511, 95% CI: 1.022–2.234, P=0.038), drinking (OR=1.706, 95% CI: 1.133–2.568, P=0.011), and tumor size ≥5 cm (OR=1.962, 95% CI: 1.248–3.084, P=0.004) subjects.
 |
Table 5 Stratification Analysis of NONHSAT053785 Expression Levels with the Risk of Intrahepatic Metastasis |
Evaluation of Serum NONHSAT053785 as a Novel Biomarker for HCC and Predictive Ability for IM
As the level of lncRNA NONHSAT053785 was markedly increased in the serum of HCC patients, we sought to determine the potential utility of serum NONHSAT053785 as a diagnostic biomarker of HCC. The ROC analysis was used to assess the diagnostic ability of serum NONHSAT053785 and AFP. The results showed that the AUC was 0.801 (95% CI: 0.746–0.855, P<0.0001) for NONHSAT053785 and 0.960 (95% CI: 0.938–0.982, P<0.0001) for AFP (Figure 4A). The sensitivity and specificity at the optimal cutoff were 73.2% and 75.4% for NONHSAT053785 and 88.4% and 93.9% for AFP, respectively. Moreover, since serum NONHSAT053785 had been established as an independent risk factor, we employed the ROC curve to evaluate the predictive ability of NONHSAT053785 and AFP expression for IM of HCC, and the results showed that the AUC was 0.678 (95% CI: 0.576–0.779, P=0.0015) for NONHSAT053785 and 0.543 (95% CI: 0.432–0.655, P>0.05) for AFP (Figure 4B). The sensitivity and specificity at the optimal cutoff were 68.2% and 64.7% for NONHSAT053785 and 40.9% and 75.0% for AFP, respectively.
Discussion
Serum tumor markers are an attractive alternative for monitoring and early diagnosis of HCC since they allow a non-invasive, objective, and reproducible assessment.17 In the present study, we explored the serum level of lncRNA NONHSAT053785 due to its potential as a novel biomarker of HCC and found that it was significantly increased in HCC patients compared to CHB and HC groups. Next, we investigated the diagnostic potential of NONHSAT053785 by calculating AUC values for both serum NONHSAT053785 and AFP. Interestingly, the AUC of AFP (0.960) in our study was much higher than the previously reported range (AUC: 0.64–0.77).18–20 This phenomenon could be due to the fact that most of our patients were at the advanced stage of HCC, and therefore, presented significantly elevated levels of AFP. The ROC curve for NONHSAT053785 showed a promising AUC value of 0.801, with a sensitivity of 73.2% and a specificity of 75.4%. Although lower than the AUC of AFP, the diagnostic efficiency of NONHSAT053785 in the current study was similar to that of AFP (AUC: 0.64–0.77), des-gamma-carboxy prothrombin (DCP, AUC: 0.71), and lectin-reactive AFP (AFP-L3, AUC: 0.73) from previous studies in HCC patients,18 thereby suggesting the diagnostic value of NONHSAT053785. However, additional studies, including a larger patient population and early HCC patients, are needed to confirm the potential diagnostic value of NONHSAT053785 in HCC.
In order to further reveal the possible influence of NONHSAT053785 on HCC, the correlations between NONHSAT053785 expression in the serum of HCC and the clinical characteristics were analyzed. The results showed that the serum level of NONHSAT053785 was significantly associated with IM, Child–Pugh classification, CEA, T-BIL, AST, ALP, and monocytes. Moreover, the level of serum NONHSAT053785 was significantly upregulated in HCC patients with IM, confirmed by the Mann–Whitney U-test. In addition, Spearman’s analysis further confirmed the correlation between serum NONHSAT053785 and clinical features. The expression of serum NONHSAT053785 was positively correlated with IM, BCLC staging, CEA, ferritin, T-BIL, AST, ALP, γ-GGT, LDH, 5ʹ-NT, WBCs, and monocytes. Together, the expression of serum NONHSAT053785 was significantly correlated with IM, tumor biomarkers, liver metabolic enzymes, and hematocytes of peripheral blood, which was confirmed by two different statistical methods. Surprisingly, these results partially corroborate with our previous research. The lncRNA NONHSAT053785 was a novel lncRNA discovered by microarray and validated through qRT-PCR in our previous study; it was found to be dysregulated in tissues of HCC patients. The previous bioinformatics analysis also verified that NONHSAT053785 was related to the biological metabolism processes. Moreover, the lncRNA-mRNA coexpression network showed that 26/28 mRNAs coexpressing with NONHSAT053785 were metabolic enzymes.14 Taken together, our previous and current results strongly suggest that NONHSAT053785 is involved in the biological metabolism processes of HCC tumorigenesis and might affect the invasion ability of the tumor. One possible hypothesis is that lncRNA NONHSAT053785 may interact with metabolism-related enzymes to promote the invasion and metastasis of HCC cells. Recent studies have shown that some circulating lncRNAs were correlated with and may be derived from peripheral blood cells. The plasma lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) expression of sepsis was positively correlated with WBC (P=0.017).21 The plasma levels of lncRNA IL-7R were correlated with WBC (P=0.0064) in patients with acute respiratory distress syndrome.22 The lncRNA CoroMarker mainly exists in the extracellular vesicles, probably from monocytes, and is stable in plasma.23 These findings were consistent with the current results that serum NONHSAT053785 expression was correlated with peripheral blood cells.
Despite advances in the technology for diagnosis and treatment, HCC often recurs locally and distantly. A common manifestation is IM, which heralds poor patient survival.24 In HCC, early recurrence due to IM leads to life-threatening tumor progression after curative surgery.25 Therefore, predicting recurrence due to IM needs to be addressed urgently with respect to HCC treatment. In addition, identifying other indicators of HCC progression is crucial. Therefore, univariate and multivariate analyses were performed to reveal the risk factors of IM. Consistent with the previous study, the current study also identified tumor size (≥5 cm) as an independent risk factor of IM.26 Notably, the upregulated serum lncRNA NONHSAT053785 has been shown to be an independent risk factor of IM. Stratified analysis deciphered that the increased risk of IM associated with the serum levels of lncRNA NONHSAT053785 was prominent in subgroups of elderly, non-smoking, drinking, and large size tumor (≥5 cm) subjects. Subsequently, the ROC curve was performed to estimate the predictive ability of NONHSAT053785 and AFP expression for IM, and the AUC was 0.678 (95% CI: 0.576–0.779, P=0.0015) for NONHSAT053785 and 0.543 (95% CI: 0.432–0.655, P>0.05) for AFP. The AUC of AFP was not statistically significant in our results, suggesting that the serum levels of lncRNA NONHSAT053785 could predict IM. Therefore, the high expression of NONHSAT053785 was identified as an independent risk factor for predicting IM in HCC. These findings suggested that NONHSAT053785 could be a potential target for developing novel therapeutic strategies for the clinical management of HCC.
To the best of our knowledge, the expression of serum lncRNA NONHSAT053785 has not been studied previously. The current results showed that serum NONHSAT053785 was positively correlated with CEA, ferritin, T-BIL, AST, ALP, γ-GGT, LDH, and 5ʹ-NT, suggesting that NONHSAT053785 may be related to cellular damage and intracellular leakage, thereby deeming it as a potential biomarker for liver damage. Moreover, it is unknown whether there are other lncRNAs that leak out into the blood stream upon cellular damage accompanying HCC and eventually increased in the serum of HCC patients. The present study mainly focused on the correlation between serum expression level of NONHSAT053785 and HCC and IM. In the subsequent studies, we will focus on the role of NONHSAT053785 in cellular damage and explore whether there are other liver-specific lncRNAs that leaks out into the blood stream upon cellular damage accompanying HCC. Notably, NONHSAT053785 is almost liver-specific.16 Therefore, the specificity of NONHSAT053785 in the diagnosis of HCC and the evaluation of liver damage is superior to some non-liver specific lncRNAs and general enzyme chemistry. However, since NONHSAT053785 in serum might not be abundant, the level needs to be detected by highly sensitive methods, such as ddPCR. Nonetheless, these limitations do not prevent the molecule from being a promising candidate as a diagnostic and predictive biomarker of HCC and IM.
Nevertheless, the present study had two limitations. First, because the majority of HCC patients were elderly men, matching the control groups and matching the gender and age in the three groups of participants was rather challenging and could not be accomplished. However, the expression of lncRNA NONHSAT053785 in HCC patients was independent of gender and age. Therefore, gender and age may have little effect on the expression level of NONHSAT053785 in the CHB and HC groups and ROC curve. Second, the mechanism of NONHSAT053785 in the oncogenesis and development of HCC and IM was not clarified. However, this residual problem is beyond the scope of the present study and would be explored in our future work.
In conclusion, the current study demonstrated that lncRNA NONHSAT053785 was an independent risk factor for IM of HCC, which was closely correlated with peripheral blood indicators and metabolic-related enzymes. The serum lncRNA NONHSAT053785 had a predictive ability for IM and may have a diagnostic value for HCC. Taken together, our results implied that NONHSAT053785 may be a crucial participant of HCC pathogenesis via tumor invasion with unknown mechanisms. These findings lay the foundation for future research on the mechanism of NONHSAT053785 in HCC. As a future research topic, one plausible hypothesis could be that lncRNA NONHSAT053785 may interact with metabolism-related enzymes to affect the tumorigenesis of HCC. Nevertheless, it needs to be confirmed by subsequent experiments. Overall, lncRNAs represent an emerging field of cancer research, and we are just beginning to understand the importance and complicity of the ncRNAs in liver carcinogenesis.7
Abbreviations
HCC, hepatocellular carcinoma; CHB, chronic hepatitis B; HC, healthy controls; ddPCR, droplet digital PCR; IM, intrahepatic metastasis; ROC, receiver operating characteristic; AUC, area under ROC curve; BCLC, Barcelona Clinic Liver Cancer; RFA, radiofrequency ablation; TACE, transarterial chemoembolization; LncRNAs, long noncoding RNAs; UCA1, urothelial carcinoma associated 1; qRT-PCR, quantitative real-time PCR; HBV, hepatitis B virus; AFP, alpha-fetoprotein; CEA, carcinoembryonic antigen; MVI, microvascular invasion; GVI, gross vascular invasion; TNM, tumor, node, metastasis; OR, odds ratio; CI, confidence interval; TP, total protein; ALB, albumin; T-BIL, total bilirubin; AST, aspartate transaminase; ALT, alanine transaminase; WBC, white blood cells; PLT, platelet; ALP, alkaline phosphatase; γ-GGT, γ-glutamyl transferase; AFU, a-L-fucosidase; CHE, cholinesterase; GLU, glucose; LDH, lactate dehydrogenase; 5ʹ-NT, 5ʹ-nucleotidase; TC, total cholesterol; TG, total triglyceride; AFP-L3, lectin-reactive AFP; DCP, des-gamma-carboxy prothrombin; MALAT1, metastasis-associated lung adenocarcinoma transcript 1.
Acknowledgments
This work was supported by the excellent talents fund of Daping Hospital and the Major Project of Army Medical University (2018 XLC 2027).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kanwal F, Singal AG. Surveillance for hepatocellular carcinoma: current best practice and future direction. Gastroenterology. 2019;157(1):54–64. doi:10.1053/j.gastro.2019.02.049
2. Yamamoto S, Midorikawa Y, Nagae G, et al. Spatial and temporal expansion of intrahepatic metastasis by molecularly-defined clonality in multiple liver cancers. Cancer Sci. 2020;111(2):601–609. doi:10.1111/cas.14282
3. Furuta M, Ueno M, Fujimoto A, et al. Whole genome sequencing discriminates hepatocellular carcinoma with intrahepatic metastasis from multi-centric tumors. J Hepatol. 2017;66(2):363–373. doi:10.1016/j.jhep.2016.09.021
4. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
5. Esposito R, Bosch N, Lanzós A, Polidori T, Pulido-Quetglas C, Johnson R. Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 screening. Cancer Cell. 2019;35(4):545–557. doi:10.1016/j.ccell.2019.01.019
6. Sadek KM, Lebda MA, Nasr NE, Nasr SM, El-Sayed Y. Role of lncRNAs as prognostic markers of hepatic cancer and potential therapeutic targeting by S-adenosylmethionine via inhibiting PI3K/Akt signaling pathways. Environ Sci Pollut Res Int. 2018;25(20):20057–20070. doi:10.1007/s11356-018-2179-8
7. Wong CM, Tsang FH, Ng IO. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat Rev Gastroenterol Hepatol. 2018;15(3):137–151. doi:10.1038/nrgastro.2017.169
8. Feng J, Yang G, Liu Y, et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9(18):5227–5245. doi:10.7150/thno.34273
9. Liu Y, Zhang YM, Ma FB, Pan SR, Liu BZ. Long noncoding RNA HOXA11-AS promotes gastric cancer cell proliferation and invasion via SRSF1 and functions as a biomarker in gastric cancer. World J Gastroenterol. 2019;25(22):2763–2775. doi:10.3748/wjg.v25.i22.2763
10. Sun L, Su Y, Liu X, et al. Serum and exosome long non coding RNAs as potential biomarkers for hepatocellular carcinoma. J Cancer. 2018;9(15):2631–2639. doi:10.7150/jca.24978
11. Yu J, Han J, Zhang J, et al. The long noncoding RNAs PVT1 and uc002mbe.2 in sera provide a new supplementary method for hepatocellular carcinoma diagnosis. Medicine (Baltimore). 2016;95(31):e4436. doi:10.1097/MD.0000000000004436
12. Chao Y, Zhou D. lncRNA-D16366 is a potential biomarker for diagnosis and prognosis of hepatocellular carcinoma. Med Sci Monit. 2019;25:6581–6586. doi:10.12659/MSM.915100
13. Zhang J, Zhang D, Zhao Q, Qi J, Li X, Qin C. A distinctively expressed long noncoding RNA, RP11-466I1.1, may serve as a prognostic biomarker in hepatocellular carcinoma. Cancer Med. 2018;7(7):2960–2968. doi:10.1002/cam4.1565
14. Li Y, Chen X, Huang H, et al. Identification of novel lncRNAs for detection of HBV-associated hepatocellular carcinoma. Onco Targets Ther. 2019;12:10199–10211. doi:10.2147/OTT.S230377
15. RNAcentral: the non-coding RNA sequence database [homepage on the Internet]. Homo sapiens (human) non-protein coding lnc-G6PC-1:1. Available from: https://rnacentral.org/rna/URS00008BF8E6/9606.
16. NONCODE: an integrated knowledge database dedicated to ncRNAs, especially lncRNAs [homepage on the Internet]. Available from: http://www.noncode.org/show_rna.php?id=NONHSAT053785&version=2&utd=1#.
17. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi:10.1016/S0140-6736(18)30010-2
18. Choi J, Kim GA, Han S, Lee W, Chun S, Lim YS. Longitudinal Assessment of Three Serum Biomarkers to Detect Very Early-Stage Hepatocellular Carcinoma. Hepatology. 2019;69(5):1983–1994. doi:10.1002/hep.30233
19. Zhang Z, Chen P, Xie H, Cao P. Using circulating tumor DNA as a novel biomarker to screen and diagnose hepatocellular carcinoma: A systematic review and meta-analysis. Cancer Med. 2020;9(4):1349–1364. doi:10.1002/cam4.2799
20. Cheng Y, Luo L, Zhang J, et al. Diagnostic value of different phenotype circulating tumor cells in hepatocellular carcinoma. J Gastrointest Surg. 2019;23(12):2354–2361. doi:10.1007/s11605-018-04067-y
21. Geng F, Liu W, Yu L. Potential role of circulating long noncoding RNA MALAT1 in predicting disease risk, severity, and patients’ survival in sepsis. J Clin Lab Anal. 2019;33(8):e22968. doi:10.1002/jcla.22968
22. Wan B, Xu WJ, Xu WN, et al. Plasma long noncoding RNA IL-7R as a prognostic biomarker for clinical outcomes in patients with acute respiratory distress syndrome. Clin Respir J. 2018;12(4):1607–1614. doi:10.1111/crj.12717
23. Yang Y, Cai Y, Wu G, et al. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin Sci (Lond). 2015;129(8):675–685. doi:10.1042/CS20150121
24. Wang Y, Xie BH, Lin WH, et al. Amplification of SMYD3 promotes tumorigenicity and intrahepatic metastasis of hepatocellular carcinoma via upregulation of CDK2 and MMP2. Oncogene. 2019;38(25):4948–4961. doi:10.1038/s41388-019-0766-x
25. Umezaki N, Nakagawa S, Yamashita YI, et al. Lysyl oxidase induces epithelial-mesenchymal transition and predicts intrahepatic metastasis of hepatocellular carcinoma. Cancer Sci. 2019;110(6):2033–2043. doi:10.1111/cas.14010
26. Hao S, Fan P, Chen S, Tu C, Wan C. Distinct recurrence risk factors for intrahepatic metastasis and multicenter occurrence after surgery in patients with hepatocellular carcinoma. J Gastrointest Surg. 2017;21(2):312–320. doi:10.1007/s11605-016-3311-z
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




