Back to Journals » Cancer Management and Research » Volume 11
A novel LARCassigner3 classification predicts outcomes in patients with locally advanced rectal cancer treated with neoadjuvant chemoradiotherapy: a retrospective training and validation analysis
Authors Zhang J, Shen L, Deng Y, Sun X, Wang Y, Yao Y, Zhang H , Zou W , Zhang Z, Wan J, Yang L, Zhu J, Zhang Z
Received 1 December 2018
Accepted for publication 19 March 2019
Published 7 May 2019 Volume 2019:11 Pages 4153—4170
DOI https://doi.org/10.2147/CMAR.S196662
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Chien-Feng Li
Jing Zhang,1,2,* Lijun Shen,1,2,* Yun Deng,1,2 Xiaoyang Sun,1,2 Yaqi Wang,1,2 Ye Yao,1,2 Hui Zhang,1,2 Wei Zou,1,2 Zhiyuan Zhang,1,2 Juefeng Wan,1,2 Lifeng Yang,1,2 Ji Zhu,1,2 Zhen Zhang1,2
1Department of Radiation Oncology, Fudan University Shanghai Cancer Center, Shanghai 200032, People’s Republic of China; 2Department of Oncology, Shanghai Medical College, Fudan University, Shanghai 200032, People’s Republic of China
*These authors contributed equally to this work
Purpose: To build and validate a predictive model of outcome for patients with locally advanced rectal cancer (LARC) treated with neoadjuvant chemoradiotherapy.
Materials and methods: We developed a LARCassigner3 classifier based on tumor and paired normal tissues of patients treated with neoadjuvant chemoradiation and surgery from January 2007 to December 2012 in Fudan University Shanghai Cancer Center. Excluding 23 pairs of tissues failed in the RNA quality test, rested 197 patients were divided into discovery (n=98) and validation (n=99) cohorts randomly. Median follow-up time was 58 months. We used the Kaplan–Meier method to estimate disease-free survival (DFS), overall survival (OS), local recurrent, and distant metastatic rate We constructed a multivariate Cox model to identify the variables independently associated with progression-free and OS.
Results: We identified three classifier genes related to relevant colorectal cancer features (CXCL9, SFRP2, and CD44) that formed the LARCassigner3 classifier assay. In the discovery set, the median DFS was 48.1 months (95% confidence interval (CI) 47.3–49.5) in the low-risk group and 23.4 months (95% CI 22.1–24.8) in the high-risk group (p=0.0134); the median OS was 39.2 months (95% CI 38.4–40.3) in the high-risk group and 19.1 months (95% CI 18.3–20.7) in the low-risk group (p=0.0134); 5-year distant metastasis was 13.9% (95% CI 9.0–21.3) in the low-risk group and 49.8% (95% CI 38.7–60.9) in the high-risk group (p=0.0072). Additionally, the different responses to neoadjuvant chemoradiotherapy and the LARCassigner3 low-risk and high-risk groups was statistically significant (p=0.004) in the discovery cohort. Similar results were obtained in the internal evaluation cohort.
Conclusions: Patients with LARCassigner3 low-risk tumors were associated with a good prognosis. The clinical utility of using LARCassigner3 subtyping for the identification of patients for neoadjuvant chemoradiotherapy requires validation in dependent clinical trial cohorts.
Keywords: prognostic marker, locally advanced rectal cancer, neoadjuvant chemoradiotherapy
Introduction
Currently, locally advanced rectal cancer (LARC) is mainly treated by neoadjuvant chemoradiotherapy (CRT) combined with total mesorectal excision (TME), which can promote regional control and maximally preserve the sphincter.1–3 As reported in some studies, superb neoadjuvant treatment response contributes to yielding satisfactory outcomes.4–6 However, neoadjuvant treatment is also associated with certain major problems, such as overtreatment and disease progression in the treatment process.7–9 Besides, individuals have different neoadjuvant CRT responses, making it difficult to be predicted. These findings suggest that a predictive test is needed to identify the factors of preoperative CRC response, which has provided greater certainties in therapeutic outcomes to LARC patients.
A great amount of tumor genomic data can be acquired thanks to the emergence of high-throughput techniques, which can thereby be comprehensively analyzed to examine the complex biological phenotypes of cancer. Importantly, biomarkers are also developed, which have been utilized to distinguish patients with specific cancer phenotypes of interest; for instance, genomic tests that have clinical utility to predict the benefit of chemotherapy to breast cancer patients have been validated and commercialized.10–12 Recently, some studies also develop the genome signatures using the unsupervised clustering approach, which are then used to classify different subtypes of colorectal cancer (CRC), and each subtype is shown to be different in terms of molecular characteristics and prognosis.13–15 Besides, different cluster numbers are recognized in different studies, which may be potentially ascribed to the heterogeneities of methods and training datasets used. For instance, the CRC classifier CRC Assigner (CRCA) has divided CRC into five distinct subtypes, including goblet-like, enterocyte, stem-like, inflammatory, as well as transit-amplifying (TA),14 and different subtypes are different in terms of prognosis. In particular, in spite of the fluorouracil-based chemotherapy, the stem cell-like subtype has a great risk of relapse;13 while the enterocyte subtype patients may greatly benefit from oxaliplatin clinically.16 As shown in studies, molecular subtypes can partly predict the benefit from chemotherapy and prognosis for colon cancer; however, the absence of independent and relevant LARC patient cohorts, the molecular subtypes that can be used for assessing individual risks and predicting the benefit of neoadjuvant CRT have not been validated yet since the related independent LARC patient cohort is lacking.
Also, the high costs of these genome-wide transcription-based methods have also severely hampered their widespread application in clinic. Several studies have suggested that the CRC molecular subtypes can be recognized using the immunohistochemical assay, which can reveal the therapeutic benefit of a specific subtype.17–19 Moreover, reverse transcriptase polymerase chain reaction (RT-PCR) accounts for another complimentary method. It is ideal to adapt the molecular tags to a small number of markers assessed by RT-PCR for the sake of patient stratification before genomic analysis becomes more common in clinical practice, which can be attributed to its cost-effectiveness and rapid implementation.20 Furthermore, the molecular characteristics governing the outcomes of LARC patients within each subtype can be further understood by analyzing the cohort after neoadjuvant CRT, respectively, in the presence of fresh-frozen operative biopsies.
On this account, the current retrospective study was carried out aiming to develop and verify a polymerase chain reaction (PCR)-based assay for patient stratification to predict the outcomes for LARC patients.
Materials and methods
Cases and samples
In the current retrospective study, altogether 220 consecutive LARC patients receiving neoadjuvant CRT and radical operation from January 2007 to December 2012 had been collected from our database that was prospectively preserved. In addition, the real-time PCR assay-based single-patient classifiers were designed by the matched pairs of frozen tissues from LARC patients that were stored at −80℃ and collected from Fudan University Shanghai Cancer Center (FUSCC) Tumor Bank. Patients conforming to the following inclusion criteria could be enrolled: 1) those with adenocarcinomas that were confirmed histologically; 2) those whose tumors were located within 12 cm above the anal verge; and 3) those at cT3-4 and/or cN1-2 stage confirmed by magnetic resonance imaging (MRI). All the patients were clinically staged with high-resolution MRI of the pelvis. The primary staging MRI was performed before CRT. Patients were imaged in a 3.0 Tesla (T) MRI (Signa Horizon, GE Medical Systems, Milwaukee, WI) using a phased-array body coil. The standard imaging protocol consisted of a sagittal T2 weighted (T2W) fast spin echo and oblique axial thin-section T2W.
Moreover, patients satisfying the following exclusion criteria should be excluded from this study: 1) those with evidence of distant metastasis (DM); 2) those with previous or simultaneous malignancies; 3) those who did not complete neoadjuvant CRT; 4) those treated by local excision or the “watch and wait” approach; 5) those who had received surgery after radiation at an interval of >16 weeks; and 6) those with inadequate tumor tissue (<5% of overall tissue sample) based on histopathological analysis.
All the enrolled patients had received conventional radiotherapy combined with the concurrent 5-fluorouracil-based chemotherapy, at the average radiation dose of 50 Gy (range, 45–55 Gy) and the daily fraction of 1.8–2.0 Gy. Besides, the XELOX regimen was adopted, and 1–3 cycles of chemotherapy were carried out at 2 weeks following the completion of radiotherapy. Rectal cancer operation was carried out 6–14 weeks following the end of neoadjuvant CRT in accordance with the principle of TME. Additionally, the XELOX or FOLFOX regimen was adopted as the postoperative adjuvant chemotherapy regimen. In the current retrospective study, the stage of patients was determined based on the Cancer Staging Manual (eighth edition) from the American Joint Committee on Cancer (AJCC). The use of the acquired tissues was approved by the Institutional Review Board (IRB) of FUSCC with informed written consents, and the protocol was approved by the hospital’s Medical Ethics Committee (IRB081280-6).
Extraction of total RNA and RT-qPCR
All the tissues tested were frozen tissue taken prior to any treatment. The total RNA was extracted from 220 pairs of LARC tissues and adjacent normal tissues using RNAiso Plus (Takara, Japan) according to the protocol of the manufacturer. Afterwards, the RNA was reversely transcribed, followed by RT-qPCR in the 96-well plates using the 7900HT Fast Real-Time PCR System (Applied Biosystems, USA) with SYBR Premix Ex Taq II (Takara, Japan). Later, ACTB (β-actin), which served as an internal reference, was used to normalize the expression of 30 candidate genes. Typically, for the extracted RNA, the A260: A280 ratio should be >1·8, while the A260: A230 ratio should be >1·6 for the sake of quality control. Specifically, 23 pairs of samples were excluded due to quality control, inadequate RNA (<0.5 μg), or the presence of a weak RT-PCR signal of ACTB (the mean cycle threshold of the reference genes, which was >35). Additionally, the expression of all genes was calculated by the 2−△△CTtumor/2−△△CTnormol approach (−△△CT=−[△CT ACTB RNA –△CT mRNA]). The primers of candidate genes are presented in Table S1.
Development and verification of the LARCassigner-3 classification
To examine whether the molecular subtype could predict the benefit of neoadjuvant treatment, the whole dataset (n=197; 23 were excluded based on RNA quality control check) was randomly divided into the discovery and verification cohorts. Moreover, a multi-step strategy was utilized to develop and verify the classifier based on gene expression, so as to stratify LARC patients in accordance with the risk factors and identify those who might possibly benefit from neoadjuvant CRT. Typically, the multi-step strategy consisted of recognizing a feature set of 30 candidate genes with high PAM scores14 and characteristics of the CRCA classifier which categorized CRC into five distinct subtypes: enterocyte, goblet-like, inflammatory, stem-like, and transit amplifying (TA) (Table 2),14,16 screening the genes that were compatible with clinical assays to identify the single-patient classifiers related to the stratification of LARC patients (discovery study), and evaluating the clinical effectiveness of the as-developed single-patient classifiers in verification cohort samples (verification study).
 | Table 1 Clinical characteristics of patients in the discovery and validation cohorts |
 | Table 2 Panel of 30 Genes related to five molecular subtypes |
Moreover, the real-time RT-PCR assay was developed an exploratory study, so as to select 30 identified candidate genes with related biological functions according to the five subtypes (goblet-like, enterocyte, stem-like, inflammatory, and TA) in LARC. On the other hand, the Cox proportional hazard regression models were also utilized to recognize the candidate genes of single-patient classifiers according to their relationships with CRT response. Then, the best threshold of normalized LARCassigner3 classifier was screened by the X-tile plots in accordance with their relationships with neoadjuvant CRT response. Typically, the X-tile plots, which were generated using the X-tile software (version 3.6.1, Yale University School of Medicine, USA), could easily and intuitively estimate the relationship of variables with CRT. Finally, the prognostic significance of LARCassigner3 was verified in the verification cohort of LARC patients.
Histopathological assessment
Pathologic TRG scores were rated for the surgically resected specimens according to the seventh guidelines of the AJCC. Based on the classification criteria, TRG 0 represents the pathological complete response (pCR), TRG 1 (moderate response) indicates the presence of single cells or cancer cells at small groups, TRG 2 (minimal response) is indicative of the outgrown residual cancer by means of fibrosis, while TRG 3 (poor response) represents the minimal or even no tumor removal and a wide residual cancer can be observed. In this study, TRG 0–1 were defined as responders, and TRG 2–3 were defined as non-responders.
Statistical analysis
The balance of clinical parameters in LARC samples collected between discovery and verification cohorts was evaluated using the chi-square test. The patient survival was counted as the duration from the date of operation to the date of events of interest or the final follow-up. Typically, three events had been determined in this study, including local recurrence, DM, and death. Data from patients that were lost to follow-up, death from all causes apart from rectal cancer, or those with no event of interest in the final follow-up were censored. Then, the survival curves were plotted according to the Kaplan–Meier method, which was then compared with log-rank tests for analysis. Besides, factors predicting prognosis were identified through univariate and multivariate analyses using the Cox proportional hazard regression models. Then, factors of p<0.10 upon univariate analysis were incorporated in the multivariate analysis. Besides, the 95% confidence interval (CI) was also calculated by means of the standard normal distribution.
The two-sided p-values were reported, with the significance level being set at <0·05. Meanwhile, the SPSS statistical software package (version 25.0) and the R software (version 3.4.2) were employed for all statistical analyses.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Written informed consent was obtained from all individual participants included in the study.
Results
Clinicopathological characteristics
Between January 2007 and December 2012, 220 LARC patients were included in this retrospective analysis. Besides, 197 pairs of LARC tissues and normal samples were collected for analysis, while 23 pairs of specimens were ruled out from assays according to the results of RNA quality control test. These 197 patients were then randomly assigned (at a ratio of 1:1) to the training or verification cohort. When judging based on the baseline pelvic CT or MRI findings, 183 LARC patients (92.9%) were at stage III before neoadjuvant CRT (including 90 and 93 in the training and verification cohorts, respectively). Meanwhile, 103 patients (52.3%) had undergone interval chemotherapy between neoadjuvant radiotherapy and surgery, and 53 (54.1%) of there were in training cohort whereas 50 (50.5%) were in verification cohort. Baseline characteristics were well matched between the two cohorts (Table 1).
Development of LARCassigner-3 classification
To translate the five identified molecular subtypes (namely, goblet-like, enterocyte, stem-like, inflammatory, and TA) in clinical test, the real-time PCR assays were designed for 30 genes closely related to these five molecular subtypes in the discovery study, including six for the goblet-like subtype, seven for the enterocyte subtype, eleven for the stem-like subtype, two for the inflammatory subtype, and four for the TA subtype (Table 2). Moreover, the expression levels of these 30 genes in 98 institutionally fresh-frozen tumor tissues and paired normal tissues were detected by real-time PCR, with β-actin gene being used as the internal reference to normalize the quantitative PCR analysis.
In the discovery cohort, univariate analysis was performed to examine the differences in the expression levels of 30 genes between responders and non-responders to neoadjuvant CRT. The results had identified that the differences in the expression levels of three genes between responders (TRG 0–1) and non-responders (TRG 2–3) were statistically significant (Table S2), which were SFRP2 (HR=1.12, 95% CI=1.02–1.23, p=0.0179), CD44 (HR=1.18, 95% CI=1.07–1.31, p=0.001) and CXCL9 (HR=0.966, 95% CI=0.932–0.998, p=0.0441). Therefore, these three classifier genes were selected, among which, CXCL9 represented the immune phenotype, while SFRP2 and CD44 stood for the stem-like phenotype.
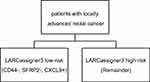
The threshold of expression for each classifier gene was established using X-tile plots, and patients were then stratified into positive or negative based on the findings of LARCassigner3 array. Ninety-eight pairs of LARC tissues and adjacent normal tissues from the FUSCC (Figure S1) were used for this analysis. Typically, the as-established thresholds for all the three genes were shown below, 7.41 for CLCX9, 3.34 for CD44, and 2.15 for SFRP2. Subsequently, the LARCassigner3 classifier was established using these three thresholds, and patients were stratified in the low-risk (high CXCL9 expression, low SFRP2, and CD44 expression), or high-risk (remainders) group in accordance with the LARCassigner3 classifier (Figure S2).
 | Figure S1 Outline of the overall study flow. Abbreviations: CRC, colorectal cancer; CRCA, CRC Assigner; FUSCC, Fudan University Shanghai Cancer Center; LARC, locally advanced rectal cancer. |
Prognostic and predictive values of LARCassigner3 classification
Afterwards, the clinical effectiveness of the LARCassigner3 classifier was tested in the discovery cohort and assessed in the verification cohort (Figure S1). Specifically, the 197 patients were followed up for a median of 58 months (IQR 5–130) to evaluate the events of interest, such as recurrence, metastasis and overall survival (OS). Among the 98 patients in the discovery cohort, the LARCassigner3 classifier had stratified 23 (23.5%) into the low-risk group, while the remaining 75 (76.5%) into the high-risk group. In our discovery set, the median OS was 48.1 (95% CI 47.3–49.5) and 23.4 months (95% CI 22.1–24.8) in the low- and high-risk groups, respectively (p=0.0134). Additionally, the median disease-free survival (DFS) was 39.2 (95% CI 38.4–40.3) and 19.1 months (95% CI 18.3–20.7) in the low- and high-risk groups, respectively (p=0.0134). Moreover, the 5-year DM rate was 13.9% (95% CI 9.0–21.3) and 49.8% (95% CI 38.7–60.9) in the low- and high-risk groups, respectively (p=0.0072) in the discovery cohort. However, no significant difference was recorded in the incidence of locoregional recurrence between these two groups (HR 0·92, 95% CI 0.79–1.08, p=0.3317, Figure 1). As for the verification cohort, the median OS was 48.1 (95% CI 47.2–49.3) and 21.8 months (95% CI 20.3–22.6) in the low- and high-risk groups, respectively (p=0.0256), while, the median DFS was 41.7 (95% CI 40.8–42.3) and 20.2 months (95% CI 19.4–21.9) in the low- and high-risk groups, respectively (p=0.0234). Moreover, the 5-year DM was 17.4% (95% CI 8.1–20.5) and 23.8% (95% CI 18.7–24.9) in the low- and high-risk groups, respectively (p=0.0296). However, no significant difference was recorded in the incidence of locoregional recurrence between these two groups (HR 0·79, 95% CI 0.70–1.07, p=0.4449, Figure 2). Therefore, the LARCassigner3 classifier high-risk group might have a worse prognosis after neoadjuvant CRT, as observed in the discovery and verification cohorts.
According to the eighth edition of the AJCC TRG scoring standard, 98 patients in the discovery cohort could be classified into responders (TRG 0–1) and non-responders (TRG 2–3). Our analysis showed that in the discovery cohort, difference in the responses to neoadjuvant CRT between the LARCassigner3 low-risk group and high-risk group was of statistical significance (p=0.004, Table 3). Similarly, the relationship between LARCassinger3 classifier and neoadjuvant CRT was verified in the verification cohort (p=0.045, Table 4), which indicated that patients in LARCassigner-3 low-risk group might have a better response to neoadjuvant CRT.
 | Table 3 LARCassigner3 classifier groups in responders (TRG 0–1) and non-responders (TRG 2–3) in the discovery cohort |
 | Table 4 LARCassigner3 classifier groups in responders (TRG 0–1) and non-responders (TRG 2–3) in the validation cohort |
We found that neither low- nor high-risk group of LARCassigner3 was correlated with age, gender, tumor stage before CRT, tumor location, chemotherapy, and surgery (Table 7). And these clinical factors were not associated with TRG groups (Table S3).
 | Table 5 Univariate analysis in relation to overall survival and disease-free survival |
 | Table 6 Multivariate analysis in relation to overall survival and disease-free survival |
 | Table 7 The relationships between clinical factors and LARCassigner3 low- or high-risk group |
Exploratory analysis of the two cohorts
Both two cohorts were used in the new exploratory analysis for OS and DFS benefits based on our plan after considering the possibility of failing the verification of LARCassigner3-RCT interaction in the absence of statistical power.
Exploratory Analyses based on the entire cohort (n=197) revealed that the median OS was 48.1 (95% CI 43.5–49.6) and 22.7 (95% CI 20.1–24.3) in the low- and high-risk groups, respectively (p=0.0225), while, the median DFS was 40.9 (95% CI 39.5–42.1) and 19.8 (95% CI 16.8–21.3) in the low- and high-risk groups, respectively (p=0.0156). Moreover, the 5-year DM rate was 15.6% (95% CI 13.2–18.9) and 31.4% (95% CI 27.8–34.3) in the low- and high-risk groups, respectively (p=0.0095; Figure 3).
The OS benefit of neoadjuvant CRT between high-risk and low-risk groups was significant upon multivariate (HR, 6.5; 95% CI, 1.97–19.5; p=0·036) and univariate analyses (HR, 8.12; 95% CI, 1.14–39.54; p=0·045; Tables 5 and 6). Besides, the 5-year DFS between low-risk and high-risk groups was significant upon univariate (HR, 8.4; 95% CI, 3.58–33.9; p=0.025) and multivariate analyses (HR, 12.63; 95% CI, 1.84–22.14; p=0.041; Tables 5 and 6).
Therefore, LARCassigner3 classifier might potentially predict the outcomes in these LARC patients. The LARCassigner3 classifier high-risk group might have a worse prognosis after neoadjuvant CRT.
Discussion
A three-gene test at clinical level was developed in the current study, which was associated with prognostic significance and could predict the response to CRT in LARC patients. Specifically, the classifier genes verified in this study were CD44, SFRP4, and CXCL9, which were screened from real-time PCR assay due to their analytical robustness and biological relevance in LARC.
Three reasons may be responsible for CXCL9 as one of the prognostic markers in rectal cancers. Firstly, TCXCL9 has been recognized to exert chemotaxis in the activated T cells as well as the NK cells;28 besides, it is also used to be an anti-cancer agent in cancer treatment research.29,30 Specifically, the anti-tumor effects of TCXCL9 are mainly regulated through accelerating the immune activities; for instance, it can elevate the responses of cytotoxic T lymphocytes, together with CD4+ or CD8+ lymphocyte infiltration into cancers, thus resisting cancer cells.31,32 Moreover, high CXCL9 and CXCL10 expression in CRC have been identified in studies to be related to the increased infiltration of memory CD8+ T cells, CD4+ T cells, as well as macrophages; besides, it is also correlated with the better prognosis and prolonged DFS.33,34 Secondly, CXCL9 can resist against angiogenesis through directly interacting with endothelium and/or recruiting T lymphocytes or NK cells, when it is used in combination with the receptor CXCR3, thus destroying tumor vasculature.35 According to the research by Ruehlmann et al, the gene therapy of CXCL9 chemokine combined with the low but not curative dose of huKS1/4-IL-2 could effectively suppress colon carcinoma growth in mice in the meantime of suppressing the pulmonary metastasis.29 Thirdly, CXCL9 and/or CXCR3 antagonism were reported to inhibit the cancer metastasis in melanoma and breast cancers also account for better prognosis.36 These dates suggest that high expression of CXCL9 may represent a useful predictor of better clinical outcome in CRC patients.
CRC patients with up-regulated STRPs have worse survival after adjuvant therapy, suggesting its vital role in chemoresistance.37 SFRPs expression has been shown to be affected by epigenetic silencing.38,39 As a WNT signaling-associated EMT modulator, SFRP2 is down-regulated in various malignancies due to the promoter hypermethylation.21 Typically, the methylation status of SFRP2 gene is a tumor-specific epigenetic marker in human breast cancer and CRC,22,23 which may play a central role in the chemoprevention of CRC.24 Besides, it is also the critical paracrine factor released by the AKT-mesenchymal stem, which can mediate myocardial survival and repair.25 These results indicate that low SFRP2 expression may contribute to the favorable prognosis for rectal cancer.
CD44 can be one of the stem cell markers for CRC,26 which can be used to be the independent factor to negatively affect the survival time for CRC patients.40–42 Mechanically, the gene expression of CD44 will be regulated through the canonical WNT signaling pathway, and it can be detected in 37% CRC patients.43,44 In addition, CRC stem cells that have high CD44 expression are associated with high invasion and metastasis capabilities.45 Generally, the 5-year survivals for CRC patients that have or have no CD44 positive cells in the primary tumor are 52.78% and 80.95%, respectively.46 The above evidence suggests that high CD44 expression indicates the poor survival of CRC.
In the current study, tumor specimens were collected from FUSCC, so as to verify whether the LARCassigner3 classifier (in which CXCL9 represented the immune phenotype, while SFRP2 and CD44 stood for the stem-like phenotype) could predict the prognosis in LARC patients. Specifically, in the discovery cohort, patients in the LARCassigner3 low-risk group had improved OS following CRT compared with those in LARCassigner3 high-risk group, and the average absolute increase in 5-year OS was 28.8%. Similar results could be discovered in the verification cohort and the whole cohort.
There is a multitude of studies to correlate gene expression data with therapy response and prognosis for LARC patients. Ghadimi et al, found in 23 LARC patients based on microarray profiling that, 54 genes had markedly different expression profiles in responders compared with the non-responders (p<0.001).47 Watanabe et al, discovered in 52 rectal cancer patients that, high expression of apoptosis inducers (including lumican, thrombospondin 2, as well as galectin-1) could be detected in responders; by contrast, high expression of apoptosis inhibitors (including cyclophilin 40 as well as glutathione peroxidase) could be found in non-responders.48 Similarly, a low predictive power level could be found when detecting the above-mentioned genes among the different cohort.47 Hur et al, found based on RT-PCR that, rectal patients with low p53 expression and/or high p21, Ki67 and CD133 expression were associated with a markedly higher tumor regression grade response as well as a higher pCR rate.49 Although previous studies have also developed factors to predict the response to CRT and the prognosis for LARC patients using the gene expression profile data, some patients are limited in terms of the reproducibility as well as the capacity to offer quantitative data. Besides, the small sample sizes in these studies may not fully represent the possible heterogeneity among LARC patients; in addition, the test as well as validation sets are lacking in these studies. Therefore, verification of our findings allowed for the development of LARCassigner3 classifier with biological value and clinical significance for LARC patients after neoadjuvant CRT. Besides, LARCassigner3 classifier assay was performed on the basis of real-time PCR, which was usually regarded as the standard RNA quantification method that can be easily performed in clinic translation.
Clinically, LARCassigner3 classifier assay, which could provide auxiliary information for the clinicians, was different from the traditional risk-stratifying approaches, like TNM staging and histological grade, and such assay could be useful in decision-making on the neoadjuvant treatments for LARC patients. The LARCassigner3 classifier could stratify patients into different groups with various long-term outcomes; in addition, it could inform the tumor biological characteristics, which could thereby complement the staging system based on anatomy. Furthermore, patients with no CRT benefits were classified into immune-low or stem-like group, which could potentially suggest the specific biological nature of relevant tumor cells.
Mark Lawler’s team had developed a method based on immunohistochemistry to determine the stem-like gene expression, so as to examine the prognosis for stage II/III CRC patients, with an HR of 2.91 (95% CI: 0.93–9.16).17 Recent research suggests that the subtypes of CMS2 and CMS3 have been subject to greater immune suppression than that of CMS1 and CMS4;50 besides, the survival for CMS2 patients is better, while that for the CMS1 cases is poor after recurrence or treatment for recurrence.43 Typically, the stem-like or immune-low CRC is found to be correlated with poor response to chemotherapy only, but not to CRT in rectal cancer. Our findings indicate that the low-risk group (high CXCL9 expression, low SFRP2 and CD44 expression) is associated with good prognosis, suggesting that stem-like or immune-low LARC may be more resistant to a long-course CRT. .
Nevertheless, our study has inevitably associated with certain limitations. Firstly, both discovery and verification analyses were performed in the absence of a preset cohort and protocol. Secondly, clinical variables recognized to influence the outcomes of patients had been well matched between discovery (n=98) and verification cohorts (n=97), but cohort specimen collection could introduce potential selection bias. Thirdly, the LARCassigner3 classifier was verified in the cohort that mostly included the Chinese LARC patients; therefore, verification in non-Chinese patients should be performed to confirm the generalizability of our findings. Moreover, verification should be performed in prospective and well-designed studies that had incorporated other races, regions, or interventions (such as, irinotecan-based CRT regimens),51,52 so as to prove the reproducibility of our findings as well as the generalizability of LARCasssigner3 classifier. In addition, the test applicability under preoperative or perioperative setting might be assessed in prospective, well designed and multi-country trials.53,54 Hopefully, the LARCassigner3 classifier proposed in the current study would be useful under other related clinical settings; for instance, in decision-making on neoadjuvant treatment and “watch and wait” strategy for LARC patients, in the presence of endoscopic biopsy samples.55,56
Conclusions
The LARCassigner3 low-risk group (high CXCL9 expression, low SFRP2 and CD44 expression) is associated with good prognosis in LARC. The LARCassigner3 classifier established in the current study to stratify LARC patients into low- and high-risk groups can potentially predict the prognosis after neoadjuvant CRT, but these results should be further verified in prospective studies.
Acknowledgments
This study was partially supported by grants from National Natural Science Foundation of China (No. 81773357, No. 81372432 and No. 81572955).
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sauer R, Liersch T, Merkel S, et al. Preoperative versus postoperative chemoradiotherapy for locally advanced rectal cancer: results of the German CAO/ARO/AIO-94 randomized phase III trial after a median follow-up of 11 years. J Clin Oncol. 2012;30(16):1926–1933. doi:10.1200/JCO.2011.40.1836
2. Bosset J, Calais G, Mineur L, et al. Enhanced tumorocidal effect of chemotherapy with preoperative radiotherapy for rectal cancer: preliminary results–EORTC 22921. J Clin Oncol. 2005;23(24):5620–5627. doi:10.1200/JCO.2005.02.113
3. Bosset J, Collette L, Calais G, et al. Chemotherapy with preoperative radiotherapy in rectal cancer. N Engl J Med. 2006;355(11):1114–1123. doi:10.1056/NEJMoa060829
4. Guillem J, Chessin D, Cohen A, et al. Long-term oncologic outcome following preoperative combined modality therapy and total mesorectal excision of locally advanced rectal cancer. Ann Surg. 2005;241(5):
5. Fokas E, Liersch T, Fietkau R, et al. Tumor regression grading after preoperative chemoradiotherapy for locally advanced rectal carcinoma revisited: updated results of the CAO/ARO/AIO-94 trial. J Clin Oncol. 2014;32(15):1554–1562. doi:10.1200/JCO.2013.54.3769
6. Huebner M, Wolff B, Smyrk T, Aakre J, Larson D. Partial pathologic response and nodal status as most significant prognostic factors for advanced rectal cancer treated with preoperative chemoradiotherapy. World J Surg. 2012;36(3):675–683. doi:10.1007/s00268-011-1409-8
7. Nacion A, Park Y, Kim N. Contemporary management of locally advanced rectal cancer: resolving issues, controversies and shifting paradigms. Chin J Cancer Res. 2018;30(1):131–146. doi:10.21147/j.issn.1000-9604.2018.01.14
8. Roselló S, Papaccio F, Roda D, Tarazona N, Cervantes A. The role of chemotherapy in localized and locally advanced rectal cancer: a systematic revision. Cancer Treat Rev. 2018;63:156–171. doi:10.1016/j.ctrv.2018.01.001
9. Wu A, Cai Y, Li Y, et al. Pattern and management of recurrence of mid-low rectal cancer after neoadjuvant intensity-modulated radiotherapy: single-center results of 687 cases. Clin Colorectal Cancer. 2018;17:e307–e313. doi:10.1016/j.clcc.2018.01.006
10. Paik S, Tang G, Shak S, et al. Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol. 2006;24(23):3726–3734. doi:10.1200/JCO.2005.04.7985
11. Wirapati P, Sotiriou C, Kunkel S, et al. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10(4):R65. doi:10.1186/bcr2124
12. Sparano J, Gray R, Makower D, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med. 2018;379:111–121. doi:10.1056/NEJMoa1804710
13. De Sousa E, Melo F, Wang X, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. doi:10.1038/nm.3174
14. Sadanandam A, Lyssiotis C, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. doi:10.1038/nm.3175
15. Roepman P, Schlicker A, Tabernero J, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134(3):552–562. doi:10.1002/ijc.28387
16. Song N, Pogue-Geile K, Gavin P, et al. Clinical outcome from oxaliplatin treatment in stage II/III colon cancer according to intrinsic subtypes: secondary analysis of NSABP C-07/NRG oncology randomized clinical trial. JAMA Oncol. 2016;2(9):1162–1169. doi:10.1001/jamaoncol.2016.2314
17. Dunne P, McArt D, Bradley C, et al. Challenging the cancer molecular stratification dogma: intratumoral heterogeneity undermines consensus molecular subtypes and potential diagnostic value in colorectal cancer. Clin Cancer Res. 2016;22(16):4095–4104. doi:10.1158/1078-0432.CCR-16-0032
18. Trinh A, Trumpi K, De Sousa E, et al. Practical and robust identification of molecular subtypes in colorectal cancer by immunohistochemistry. Clin Cancer Res. 2017;23(2):387–398. doi:10.1158/1078-0432.CCR-16-0680
19. Trumpi K, Ubink I, Trinh A, et al. Neoadjuvant chemotherapy affects molecular classification of colorectal tumors. Oncogenesis. 2017;6(7):e357. doi:10.1038/oncsis.2017.48
20. Reed S, Dinan M, Schulman K, Lyman G. Cost-effectiveness of the 21-gene recurrence score assay in the context of multifactorial decision making to guide chemotherapy for early-stage breast cancer. Genet Med. 2013;15(3):203–211. doi:10.1038/gim.2012.119
21. Zhang X, Song Y, Lu H, et al. Combined detection of plasma GATA5 and SFRP2 methylation is a valid noninvasive biomarker for colorectal cancer and adenomas. World J Gastroenterol. 2015;21(9):2629–2637. doi:10.3748/wjg.v21.i9.2629
22. Bagci B, Sari M, Karadayi K, Turan M, Ozdemir O, Bagci G. KRAS, BRAF oncogene mutations and tissue specific promoter hypermethylation of tumor suppressor SFRP2, DAPK1, MGMT, HIC1 and p16 genes in colorectal cancer patients. Cancer Biomark. 2016;17(2):133–143. doi:10.3233/CBM-160624
23. Veeck J, Noetzel E, Bektas N, et al. Promoter hypermethylation of the SFRP2 gene is a high-frequent alteration and tumor-specific epigenetic marker in human breast cancer. Mol Cancer. 2008;7:83. doi:10.1186/1476-4598-7-83
24. Chen L, Jiang B, Zhong C, et al. Chemoprevention of colorectal cancer by black raspberry anthocyanins involved the modulation of gut microbiota and SFRP2 demethylation. Carcinogenesis. 2018;39(3):471–481. doi:10.1093/carcin/bgy009
25. Mirotsou M, Zhang Z, Deb A, et al. Secreted frizzled related protein 2 (SFRP2) is the key Akt-mesenchymal stem cell-released paracrine factor mediating myocardial survival and repair. Proc Natl Acad Sci USA. 2007;104(5):1643–1648. doi:10.1073/pnas.0610024104
26. Morath I, Hartmann T, Orian-Rousseau V. CD44: more than a mere stem cell marker. Int J Biochem Cell Biol. 2016;81(Pt A):166–173. doi:10.1016/j.biocel.2016.09.009
27. Mueller N, Wicklein D, Eisenwort G, et al. CD44 is a RAS/STAT5-regulated invasion receptor that triggers disease expansion in advanced mastocytosis. Blood. 2018;132:1936–1950. doi:10.1182/blood-2018-02-833582
28. Hertenstein A, Schumacher T, Litzenburger U, et al. Suppression of human CD4+ T cell activation by 3,4-dimethoxycinnamonyl-anthranilic acid (tranilast) is mediated by CXCL9 and CXCL10. Biochem Pharmacol. 2011;82(6):632–641. doi:10.1016/j.bcp.2011.06.013
29. Ruehlmann JM, Xiang R, Niethammer AG, et al. MIG (CXCL9) chemokine gene therapy combines with antibody-cytokine fusion protein to suppress growth and dissemination of murine colon carcinoma. Cancer Res. 2001;61(23):8498–8503.
30. Zhang R, Tian L, Chen LJ, et al. Combination of MIG (CXCL9) chemokine gene therapy with low-dose Cisplatin improves therapeutic efficacy against murine carcinoma. Gene Ther. 2006;13(17):1263–1271. doi:10.1038/sj.gt.3302756
31. Ben-Baruch A. The multifaceted roles of chemokines in malignancy. Cancer Metastasis Rev. 2006;25(3):357–371. doi:10.1007/s10555-006-9003-5
32. Li G, Tian L, Hou JM, et al. Improved therapeutic effectiveness by combining recombinant CXC chemokine ligand 10 with Cisplatin in solid tumors. Clin Cancer Res. 2005;11(11):4217–4224. doi:10.1158/1078-0432.CCR-04-2117
33. Jiang Z, Xu Y, Cai S. CXCL10 expression and prognostic significance in stage II and III colorectal cancer. Mol Biol Rep. 2010;37(6):3029–3036. doi:10.1007/s11033-009-9873-z
34. Mlecnik B, Tosolini M, Charoentong P, et al. Biomolecular network reconstruction identifies T-cell homing factors associated with survival in colorectal cancer. Gastroenterology. 2010;138(4):1429–1440. doi:10.1053/j.gastro.2009.10.057
35. Frederick MJ, Clayman GL. Chemokines in cancer. Expert Rev Mol Med. 2001;3(19):1–18. doi:10.1017/S1462399401003301
36. Kawada K, Sonoshita M, Sakashita H, et al. Pivotal role of CXCR3 in melanoma cell metastasis to lymph nodes. Cancer Res. 2004;64(11):4010–4017. doi:10.1158/0008-5472.CAN-03-1757
37. Buess M, Terracciano L, Reuter J, et al. STRAP is a strong predictive marker of adjuvant chemotherapy benefit in colorectal cancer. Neoplasia (New York, NY). 2004;6(6):813–820. doi:10.1593/neo.04307
38. Aguilera O, Muñoz A, Esteller M, Fraga MF. Epigenetic alterations of the Wnt/beta-catenin pathway in human disease. Endocr Metab Immune Disord Drug Targets. 2007;7(1):13–21. doi:10.2174/187153007780059450
39. Caldwell GM, Jones CE, Taniere P, et al. The WNT antagonist SFRP1 is downregulated in premalignant large bowel adenomas. Br J Cancer. 2006;94(6):922–927. doi:10.1038/sj.bjc.6602967
40. Zhao LH, Lin QL, Wei J, Huai YL, Wang KJ, Yan HY. CD44v6 expression in patients with stage II or stage III sporadic colorectal cancer is superior to CD44 expression for predicting progression. Int J Clin Exp Pathol. 2015;8(1):692–701.
41. Zlobec I, Terracciano L, Tornillo L, et al. Role of RHAMM within the hierarchy of well-established prognostic factors in colorectal cancer. Gut. 2008;57(10):1413–1419. doi:10.1136/gut.2007.141192
42. Garouniatis A, Zizi-Sermpetzoglou A, Rizos S, Kostakis A, Nikiteas N, Papavassiliou AG. FAK, CD44v6, c-Met and EGFR in colorectal cancer parameters: tumour progression, metastasis, patient survival and receptor crosstalk. Int J Colorectal Dis. 2013;28(1):9–18. doi:10.1007/s00384-012-1520-9
43. Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi:10.1038/nm.3967
44. Wielenga VJ, Smits R, Korinek V, et al. Expression of CD44 in APC and TCF mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154(2):515–523. doi:10.1016/S0002-9440(10)65297-2
45. Todaro M, Gaggianesi M, Catalano V, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014;14(3):342–356. doi:10.1016/j.stem.2014.01.009
46. Li XD, Ji M, Wu J, Jiang JT, Wu CP. Clinical significance of CD44 variants expression in colorectal cancer. Tumori. 2013;99(1):88–92. doi:10.1700/1248.13794
47. Ghadimi BM, Grade M, Difilippantonio MJ, et al. Effectiveness of gene expression profiling for response prediction of rectal adenocarcinomas to preoperative chemoradiotherapy. J Clin Oncol. 2005;23(9):1826–1838. doi:10.1200/JCO.2005.00.406
48. Watanabe T, Komuro Y, Kiyomatsu T, et al. Prediction of sensitivity of rectal cancer cells in response to preoperative radiotherapy by DNA microarray analysis of gene expression profiles. Cancer Res. 2006;66(7):3370–3374. doi:10.1158/0008-5472.CAN-05-3834
49. Hur H, Tulina I, Cho MS, et al. Biomarker-based scoring system for prediction of tumor response after preoperative chemoradiotherapy in rectal cancer by reverse transcriptase polymerase chain reaction analysis. Dis Colon Rectum. 2016;59(12):1174–1182. doi:10.1097/DCR.0000000000000711
50. Lal N, White BS, Goussous G, et al. KRAS mutation and consensus molecular subtypes 2 and 3 are independently associated with reduced immune infiltration and reactivity in colorectal cancer. Clin Cancer Res. 2018;24(1):224–233. doi:10.1158/1078-0432.CCR-17-1090
51. Isella C, Brundu F, Bellomo S, et al. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat Commun. 2017;8:15107. doi:10.1038/ncomms15107
52. Karpinski P, Rossowska J, Sasiadek M. Immunological landscape of consensus clusters in colorectal cancer. Oncotarget. 2017;8(62):105299–105311. doi:10.18632/oncotarget.22169
53. Sveen A, Bruun J, Eide P, et al. Colorectal cancer consensus molecular subtypes translated to preclinical models uncover potentially targetable cancer cell dependencies. Clin Cancer Res. 2018;24(4):794–806. doi:10.1158/1078-0432.CCR-17-1234
54. Alderdice M, Richman S, Gollins S, et al. Prospective patient stratification into robust cancer-cell intrinsic subtypes from colorectal cancer biopsies. J Pathol. 2018. doi:10.1002/path.5051
55. Maas M, Beets-Tan R, Lambregts D, et al. Wait-and-see policy for clinical complete responders after chemoradiation for rectal cancer. J Clin Oncol. 2011;29(35):4633–4640. doi:10.1200/JCO.2011.37.7176
56. Rullier E, Rouanet P, Tuech J, et al. Organ preservation for rectal cancer (GRECCAR 2): a prospective, randomised, open-label, multicentre, phase 3 trial. Lancet. 2017;390(10093):469–479. doi:10.1016/S0140-6736(17)31056-5
Supplementary Materials
 | Table S1 Primers used for qRT-PCR |
 | Table S2 Classifier genes selected by Cox proportional hazard regression models |
 | Table S3 The relationships between clinical factors and TRG |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.