Back to Journals » OncoTargets and Therapy » Volume 14
A Novel Circular RNA circCSPP1 Promotes Liver Cancer Progression by Sponging miR-1182
Authors Jia N, Song Z, Chen B, Cheng J, Zhou W
Received 24 November 2020
Accepted for publication 18 March 2021
Published 23 April 2021 Volume 2021:14 Pages 2829—2838
DOI https://doi.org/10.2147/OTT.S292320
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Alberto Bongiovanni
Nan Jia, Zhe Song, Baosheng Chen, Jinsheng Cheng, Wenyong Zhou
Department of General Surgery, CangZhou General Hospital, CangZhou, Hebei, 061001, People’s Republic of China
Correspondence: Nan Jia
Department of General Surgery, CangZhou General Hospital, No. 16 Xinhua West Road, CangZhou, Hebei, 061001, People’s Republic of China
Tel +86-18031792208
Email [email protected]
Introduction: Aberrant circular RNA (circRNA) expression has been extensively discovered for its involvement in both the initiation and progression of various cancers. Through screening circRNA profile, we identified a novel circRNA has_circ_0001806, which is termed as circCSPP1 in liver cancer. In the present study, we aim to investigate the role of circCSPP1 in the progression of liver cancer.
Methods: Fluorescence in situ hybridization (FISH) was used to detect the location of circCSPP1. Function studies including MTT, colony formation assay, transwell assay and flow cytometry were carried out to detect the malignant behaviour of circCSPP1 on liver cancer cells. Luciferase assay and RNA pull down were used to detect the interaction between miR-1182 and circCSPP1 as well as RAB15. Quantitative realtime (qPCR) and Western blot were performed to evaluate the RNA and protein expression, respectively.
Results: CircCSPP1 knockdown inhibited the proliferation, migration and invasion while promoted apoptosis of liver cancer cells. Mechanically, we predicted and verified the target miR of circCSPP1 which is miR-1182. miR-1182 was capable of reversing the effect of circCSPP1 on liver cancer cells. Moreover, miR-1182 was found to also target RAB15 to participate in the regulation of cell phenotype.
Discussion: Taken together, circCSPP1 promoted progression of liver cancer cells via sponging miR-1182 which may serve as a novel prognostic and therapeutic target for liver cancer.
Keywords: circRNA, circCSPP1, liver cancer, miR-1182, RAB15
Introduction
Liver cancer has become the third leading cause of cancer-associated mortality worldwide.1 The main therapeutic methods for liver cancer include radical resection and chemotherapy. The treatment of liver cancer has made significant progress in the past few years. However, the prognosis of liver cancer remains poor.2,3 The current 5-year survival rate of liver cancer is estimated to be <25% due to the frequent recurrence and metastasis.4 Therefore, it is urgent to extend our understanding of the molecular mechanisms underlying liver cancer development.
Circular RNAs (circRNAs) are a class of non-coding RNAs characterized by a covalently closed loop without 5ʹ to 3ʹ polyadenylated tails. These molecules are formed by back-splicing events.5 Currently, circRNAs have been shown to be highly conserved and are expressed with high stability across different species.6 circRNAs are differentially expressed in various types of cancer, including prostate,7 breast8 and hepatocellular9 cancer. In addition, they are also involved in several physiological and pathophysiological processes, such as adsorption of microRNAs (miRNAs/miRs), regulation of alternative splicing, protein-RNA interaction and DNA and transcription factor expression, and ultimately, alteration of the cell phenotype.10,11
miRNAs are small endogenous non-coding RNAs, which can regulate fundamental cell processes by binding to the specific 3ʹuntranslated regions of target genes.12 This binding can inhibit protein translation or induce degradation of target mRNAs. circRNAs mainly sponge miRNAs to influence gene expression and biological functions.13,14
In the present study, the circRNA circCSPP1 was identified as an upregulated circRNA in liver cancer tissues and cell lines. Loss- and gain-of-function studies were performed to determine the function of circCSPP1 in liver tumorigenesis. The data predicted and verified the target miRNA of circCSPP1, which was miR-1182, and further elucidated its underlying mechanism in liver cancer. In summary, circ-CSPP1 functioned as an oncogene in liver cancer and may be a potential therapeutic target for this disease.
Methods and Materials
Patients and Specimens
The cohort study was conducted on samples collected between January 2017 and January 2018. Patients with a confirmed diagnosis of liver cancer were included. Patients who had received previous chemotherapy, radiotherapy or biological medication were excluded from the study. A total of 55 pairs of liver cancer and normal tissues were obtained from Cangzhou Central Hospital. The distance between tumor and adjacent tissue was 2–3 cm. The tissues were frozen in liquid nitrogen immediately after excision and subsequently stored at −80°C. All experimental procedures were approved by the Ethics Committee of Cangzhou Central Hospital and were performed in accordance with the Declaration of Helsinki of the World Medical Association. Written informed consent was obtained from each patient.
Cell Culture
The liver cancer cell lines (HepG2, Huh7, MHCC97L, LM3, Hep3B) and the human liver epithelial-2 cell (THLE-2) were purchased from the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI-1640 medium (Invitrogen, Thermo Fisher Scientific, Inc., MA, USA) containing 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Inc., Australia), 100 U/mL penicillin and 100 mg/mL streptomycin (Gibco, Thermo Fisher Scientific, Inc., MA, USA). All cell lines were incubated in a humidified atmosphere of 5% CO2 at 37°C.
Cell Transfection
miR-1182 mimic and mimic control were synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). circCSPP1 overexpressing vector, RAB15 overexpressing vector, control vector, small interfering (si)RNA for circCSPP1, control siRNA and short hairpin RNA (shRNA) targeting circCSPP1 were synthesized by Hanbio Biotechnology Co., Ltd. (Beijing, China). The cells were resuspended and seeded on six-well plates at a density of 8×105 cells and cultured in RPMI-1640 without FBS at 37°C for 12 h prior to transfection. The sequences for the siRNA and mimics were as follows:
miR-1182 mimic (forward: 5ʹGAGGGUCUUGGGAGGGAUGUGAC3ʹ, reverse: 5ʹ CAGUGUAGGGAGGGUUCUGGGAG3ʹ); mimic control (forward: 5ʹGCATATTGCATGCCTTAAG3ʹ, reverse: 5ʹ CTTAAGGCATGCAATATGC3ʹ); si-nc (forward: 5ʹAATTCAAAAAGCAATGCATTTCAACCT3ʹ, reverse: 5ʹCCGGAGGTTGAAATGCATTGCTTTTTG3ʹ); si-circCSPPA (forward: 5ʹCCCAGTGCTCCAGACAATGAA3ʹ, reverse: 5ʹTTCATTGTCTGGAGCACTGGG3ʹ)
Cell Counting Kit (CCK)-8
Following transfection, the cells were collected and re-suspended in culture medium, followed by seeding into 96-well plates at a density of 1x104 cells/well. The cells were subsequently treated with 15 μL CCK-8 solution for an additional 2 h at 37°C. The absorbance was detected at 480 nm using a microplate reader (BioRad Laboratories, Inc., CA, USA). Each experiment was performed at least 3 times.
Transwell Experiment
Following transfection, 1x105 cells were suspended in 200 μL RPMI-1640 medium and were seeded on the upper Transwell chamber (Corning, Inc., NY, USA). Following 24 h of incubation at 37°C, the cells were fixed on the surfaces of lower chambers in 20% methanol followed by staining with 1% crystal violet (Bicobio, Tianjin, China). Finally, the cell colonies were imaged and counted.
Colony Formation Assay
Following transfection, the cells were seeded in 12-well plates at a density of 100 cells/well. The cells were incubated for 2 weeks, fixed in 10% formaldehyde and stained with 1% crystal violet (Beyotime Institute of Biotechnology, Shanghai, China). The images were photographed using a microscope (Leica Microsystems GmbH, Germany) and the colonies that consisted of >50 cells were counted.
Western Blot Analysis
The total protein was extracted and the concentration of the collected protein was determined in a Nanodrop 2000 system (Thermo Fisher Scientific, Inc., USA). SDS-PAGE was performed and 40 μg protein was loaded for electrophoresis, followed by transfer to a PVDF membrane. The blots were subsequently blocked in 5% skimmed milk at room temperature for 2 h, incubated with the primary antibody (1:1000) overnight at 4°C followed by further incubation with the secondary antibody (1:1000) at room temperature for an additional 2 h. Subsequently, the blots were visualized using an ECL chemiluminescence kit and the gray value of each band was analyzed with QuantityOne software.
RNA-Fluorescence in situ Hybridization
Hybridization of Alexa Fluor 555-labeled circCSPP1 was performed according to the manufacturer’s protocol (Shanghai GenePharma Co., Ltd., Shanghai, China). FISH assay was performed using a fluorescence in situ hybridization kit according to the manufacturer’s instructions (Guangzhou RiboBio Co., Ltd., Guangzhou, China). DAPI was used to stain the cell nucleus. The subcellular distribution of circCSPP1 was observed with a confocal laser scanning microscope (Olympus FV1000, Olympus Corporation, Japan).
RNA Pull-Down Assay
The biotinylated probe of miR-1182 and the control probe were obtained from Hanbio Biotechnology Co., Ltd. (Shanghai, China). The streptavidin-coated beads were initially coated with the probe. The cells were lysed to extract the total protein. Following pretreatment with magnetic beads, RNA and beads were mixed. Following washing for 5 times, the RNA in the pull-down complex was extracted using the TRIzol® reagent and analyzed by qPCR.
Dual Luciferase Reporter Gene Assay
The luciferase assay was performed using the dual-luciferase reporting system psiCHECKTM (Thermo Fisher Scientific, Inc.). CircCSPP1 or RAB15 wild-type (WT) and its mutant sequence (mut) were cloned into plasmid psiCHECK2. 293T cells (4x104 cells/well) were cultured overnight in 24-well plates. The psiCHECK2 and the Renilla luciferase expression plasmids were transfected into 293T cells using Lipofectamine 2000®. Following 24 h of cell incubation, luciferase activity was measured by the Luciferase reporter gene assay system (Promega Corporation).
Xenograft Model
HepG2 and Huh7 cells were collected and resuspended in RPMI-1640medium. A total of 3 × 106 HepG2 cells (100 μL) were injected into the posterior flank of 24 mice subcutaneously. The length (L) and width (W) of the tumors were monitored every 3 days. Following a 35-day period of growth, significant differences were identified with regard to the tumor volume between the two groups. All the mice were euthanized by intraperitoneal injection of an overdose of pentobarbital sodium (160 mg/kg). Subsequently, the tumors were removed and weighed. All animal procedures were approved by the Animal Research Committee of Cangzhou Central Hospital (approval no. CZCH-20190858). People’s Republic of China National Standard (GB/T35892-2018) was followed for the welfare of the laboratory animals.
Results
circCSPP1 is Highly Expressed in HSC Tissues and Cells
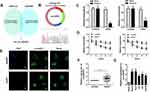
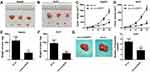
To identify the dysregulated circRNAs, the GSE97332 and GSE94508 datasets were analyzed in the GEO database using the GEO2R tool. hsa_circ_0001806 expression was dysregulated in both datasets (Figure 1A). It is well known that circRNAs are more stable due to their looped structure. First, sanger sequence was performed to check the exist of circCSPP1. As Figure 1B showed, the sequencing confirmed the existence of circCSPP1. To confirm the stability of circCSPP1, RNase R was employed in the experiments. The results indicated that, following RNase R treatment, the levels of the linear forms of CSPP1 were notably decreased, while no significant changes were observed in the levels of circCSPP1, which indicated the optimal stability of circCSPP1 (Figure 1C). Furthermore, liver cancer cells were treated with actinomycin D to inhibit transcription. The half-lives of circCSPP1 and linear CSPP1 were subsequently evaluated. The results indicated that circCSPP1 had a longer half-life than linear CSPP1 (Figure 1D). In addition, specific probes were designed to carry out FISH analysis. Figure 1E revealed that circCSPP1 was located mainly in the cytoplasm. Then, the expression levels of circCSPP1 in the cancer tissues or the paired normal tissues were assessed using qPCR. The 55 cases were divided into two groups: a low circCSPP1 group (n=31) and a high circCSPP1 group n=24). The correlation of clinical parameters from these 55 cases was analyzed and shown in Table 1. It was found that circCSPP1 expression was correlated with histological grade and tumor size (Table 1). The results demonstrated that circCSPP1 expression was markedly upregulated in liver cancer tissues and cells compared with that in normal tissues and in human liver epithelial-2 cells (Figure 1F and G).
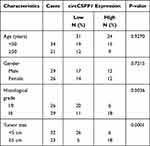 |
Table 1 Clinicopathological Characteristics of HCC Patients (n=55) |
Knockdown of circCSPP1 Inhibits the Proliferation and Migration of Liver Cancer Cells
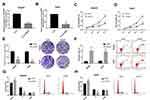
Due to the altered expression of circCSPP1 in liver cancer tissues and cell lines, it was hypothesized that circCSPP1 played a critical role in the progression of liver cancer. Loss of function studies were carried out using siRNA of circCSPP1. qPCR analysis verified the efficiency of the siRNA knockdown for circCSPP1 (Figure 2A and B). MTT results indicated that knockdown of circCSPP1 expression inhibited the proliferation of liver cancer cells (Figure 2C and D). Colony formation assay demonstrated that circCSPP1 knockdown attenuated the colony formation ability of liver cancer cells (Figure 2E). Flow cytometry was subsequently performed to evaluate the induction of apoptosis and the cell cycle distribution of liver cancer cells. The data indicated that circCSPP1 knockdown induced a significant increase in cell apoptosis (Figure 2F) and a concomitant G0/G1 arrest of liver cancer cells (Figure 2G and H). In addition, transwell assays were performed to evaluate the migratory and invasive abilities of liver cancer cells. The results indicated that circCSPP1 knockdown reduced the number of migrating (Figure 3A–C) and invading cells (Figure 3D–F).
circCSPP1 Exerts Its Effects by Sponging miR-1182
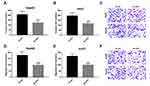
It is well known that circRNAs sponge miRNAs in order to regulate expression of downstream genes, which participate in several cell physiological processes. In the current study, bioinformatics analysis was performed using the specific software Circinteractome to predict the potential miRNA targets of circCSPP1. The cells were transfected with different types of miRNA mimics. Luciferase activity was assessed. Control mimics was transfected into the scramble mimic cell group. It was found that miR-1182 and miR-486-3p inhibited luciferase activity (Figure 4A) in the cells. Figure 4B demonstrates that miR-1182 expression was downregulated in liver cancer cells. In addition, RNA pull-down assay indicated that miR-1182 interacted with circCSPP1 directly (Figure 4C and D). The complementary base sequences between circCSPP1 and miR-1182 are shown in Figure 4E. The qPCR results indicated that the miR-1182 mimic notably promoted the expression levels of miR-1182 (Figure 4F). The luciferase activity assay was conducted to verify the binding of circCSPP1 and miR-1182. The data indicated that miR-1182 decreased the luciferase activity levels of the wild-type reporter for circCSPP1, whereas this effect was not observed with the mutant-type reporter plasmid, which confirmed that miR-1182 was a sponge target of circCSPP1 (Figure 4G).
miR-1182 Targets RAB15 in Liver Cancer Cells
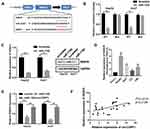
To elucidate the precise mechanism underlying the function of circCSPP1, the downstream targets of miR-1182 were investigated. Figure 5A indicates the complementary base sequences between RAB15 and miR-1182. Luciferase activity assay was conducted to verify the binding of RAB15 and miR-1182. miR-1182 decreased the luciferase activity of the WT reporter for RAB15 but not that of the MUT-type reporter, which confirmed miR-1182 as a sponge target of RAB15 (Figure 5B). Western blot analysis was used to evaluate the expression levels of RAB15 following miR-1182 overexpression and knockdown. The data indicated that miR-1182 significantly inhibited the expression levels of RAB15, while miR-1182 knockdown promoted RAB15 expression (Figure 5C). Figure 5D demonstrated that RAB15 expression levels were upregulated in liver cancer cells. qPCR results indicated that miR-1182 reduced the expression levels of RAB15, while circCSPP1 overexpression reversed the effects of miR-1182, demonstrating the competing endogenous (ce)RNA association between RAB15 and circCSPP1 (Figure 5E). Pearson’s analysis indicated a positive correlation between RAB15 and circCSPP1 (Figure 5F).
RAB15 Reverses the Function of circCSPP1 Knockdown in Liver Cancer
As a ceRNA of circCSPP1, it was predicted that RAB15 may also be involved in the biological processes of liver cancer cells. Co-transfection of si-circCSPP1 and RAB15 overexpressing vector were performed. Transfection of the cells with the RAB15 overexpressing vector caused a significant elevation in the levels of RAB15. Co-transfection of the si-circCSPP1 and the RAB15 overexpressing vector into the cells notably increased the levels of RAB15 compared to those of the si-circCSPP1 single-transfection group (Figure 6A). By using functional studies, it was observed that RAB15 overexpression reversed the effect of circCSPP1 knockdown on cell proliferation (Figure 6B and C), colony formation (Figure 6D), migration (Figure 6E) and invasion (Figure 6F).
circCSPP1 Promotes the Growth of Liver Cancer Cells in vivo
Subsequently, an in vivo study was carried out to further investigate the biological effects of circCSPP1 in the progression of liver cancer. A stable HepG2 cell line was established following transfection of circCSPP1 knockdown and control vectors. Tumor growth inhibition was induced by circCSPP1 knockdown (Figure 7A–D). The weight of the tumors was reduced by si-circCSPP1 (Figure 7E and F). The maximum tumor diameter and volume allowed in these studies is 20 mm and 1500 mm3, respectively. The maximum tumor diameter and volume observed in the present study was 17.5 mm and 1432 mm3, respectively. Furthermore, as circCSPP1 regulated the migration and invasion ability of liver cancer cells. We established tumor metastasis model. The results showed that circCSPP1 knockdown could reduce the number of metastatic nodules (Figure 7G).
Discussion
circRNAs have been shown to play crucial roles in certain cellular functions, such as proliferation, apoptosis, differentiation and metabolism.15 In the present study, a novel circRNA was identified, which was denoted by circCSPP1. The data demonstrated that circCSPP1 originated from exons 3 to 9 of CSPP1 and formed a loop structure via connecting the 3ʹ and 5ʹ splice sites. The stability of circCSPP1 was confirmed by its stable expression under RNase R digestion. To the best of our knowledge, this is the first study of circCSPP1 that focused on its altered expression and biological effects. The upregulated expression and the stability of circCSPP1 render it a potential biomarker and a diagnostic and therapeutic target. However, HepG2 used as a liver cancer cell are challenged now, this is a limitation of the present study. We will study the effect of circCSPP1 on the other liver cancer cell lines in our future work.
Various miRNAs have been shown to participate in the biological processes of liver cancer. miR-497, −320, −613, −215 and −145 are associated with decreased proliferation, migration and invasion of human liver cancer cells via targeting vascular endothelial growth factor A, specificity protein 1, liver cancer tumor suppressor gene 1 and ADAM19.16–21 Among them, miR-1182 which belongs to the miR-1182 family has been extensively studied and shown to play critical roles in cellular differentiation and development. Several studies have suggested the oncogenic or anti-cancer effect of miR-1182 in different types of cancer including ovarian cancer,22 myeloid leukemia,23 colorectal cancer,24 non-small cell lung cancer25 and gastric cancer.26 However, to the best of our knowledge, miR-1182 has not been studied in liver cancer to date. Moreover, its wide expression in human organs, notably during early ocular development, has attracted considerable attention.27 miR-1182 has been shown to target hsa_circ_007624828 as well as circRNA8073,29 and participates in the regulation of cell apoptosis and proliferation.
In the present study, miR-1182 was shown to be the target miRNA of circCSPP1, whose expression was downregulated in liver cancer tissues and cells. Moreover, functional studies revealed for the first time the anti-cancer effects of miR-1182 in liver cancer. Of note, miR-1182 overexpression reversed the effect of circCSPP1, which further verified the association between these two target RNA molecules. In order to further elucidate the underlying mechanism, the target gene of miR-1182 was predicted. The analysis verified RAB15 as a potential target gene. RAB15 expression is closely associated with the susceptibility of cells to DNA damage-induced cell death. This feature may be involved in the regulation of the liver cancer cell phenotype. In future studies, the effects of RAB15 on DNA damage and the associated molecular mechanism involving circCSPP1 and miR-1182 will be investigated.
Insulin-resistant livers may coordinate impaired hepatic metabolic function and increase the risk of liver cancer. miR-833b and miR-205 have been reported as the potential cooperative modulators of liver function.30 These findings indicated the role of miR-1182 in liver metabolism, which correlated with the progression of liver cancer. This research content is worth studying. In addition, the function of circCSPP1 in HepG2 and Huh7 cells was explored. HLF or HLE hepatoma cell lines exhibit an aggressive malignant phenotype and rapid progression. HLF or HLE cell lines will be investigated in future studies to provide further support for the current conclusions. This is one of the limitations of the present study. The mechanism underlying the effects of circCSPP1 in liver cancer will be further assessed in future studies.
In conclusion, the findings of the present study revealed that circCSPP1 promoted the malignant behavior of liver cancer by sponging miR-1182, suggesting its potential as a diagnostic or therapeutic target in liver cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J and Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Liu Z, Yu Y, Huang Z, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12):900. doi:10.1038/s41419-019-2089-9
3. Zhu K, Zhan H, Peng Y, et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. 2019.
4. El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi:10.1056/NEJMra1001683
5. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881. doi:10.1016/j.cell.2018.12.021
6. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi:10.1002/hep.29270
7. Chen S, Huang V, Xu X, et al. Widespread and functional RNA circularization in localized prostate cancer. Cell. 2019;176(4):831–843. doi:10.1016/j.cell.2019.01.025
8. Smid M, Wilting SM, Uhr K, et al. The circular RNome of primary breast cancer. Genome Res. 2019;29(3):356–366. doi:10.1101/gr.238121.118
9. Yu J, Xu QG, Wang ZG, et al. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J Hepatol. 2018;68(6):1214–1227. doi:10.1016/j.jhep.2018.01.012
10. Hu ZQ, Zhou SL, Li J, et al. Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. 2019;70(4):1214–1230. doi:10.1002/hep.30630
11. Chien Y, Tsai PH, Lai YH, et al. CircularRNA as novel biomarkers in liver diseases. J Chin Med Assoc. 2019.
12. Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. 2019;16(8):503–514. doi:10.1038/s41569-019-0185-2
13. Bezzi M, Guarnerio J, Pandolfi PP. A circular twist on microRNA regulation. Cell Res. 2017;27(12):1401–1402. doi:10.1038/cr.2017.136
14. Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43(4):2454–2465. doi:10.1093/nar/gkv045
15. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
16. Sun Z, Zhang A, Jiang T, Du Z, Che C, Wang F. MiR-145 suppressed human retinoblastoma cell proliferation and invasion by targeting ADAM19. Int J Clin Exp Pathol. 2015;8(11):14521–14527.
17. Li J, Zhang Y, Wang X, Zhao R. microRNA-497 overexpression decreases proliferation, migration and invasion of human retinoblastoma cells via targeting vascular endothelial growth factor A. Oncol Lett. 2017;13(6):5021–5027. doi:10.3892/ol.2017.6083
18. Zhao Y, Zhang S, Zhang Y. MicroRNA-320 inhibits cell proliferation, migration and invasion in retinoblastoma by targeting specificity protein 1. Mol Med Rep. 2017;16(2):2191–2198. doi:10.3892/mmr.2017.6767
19. Zhang Y, Zhu X, Zhu X, et al. MiR-613 suppresses retinoblastoma cell proliferation, invasion, and tumor formation by targeting E2F5. Tumour Biol. 2017;39:1393397990.
20. Chen Z, Liu K, Li L, Chen Y, Du S. miR-215 promotes cell migration and invasion of gastric cancer by targeting Retinoblastoma tumor suppressor gene 1. Pathol Res Pract. 2017;213(8):889–894. doi:10.1016/j.prp.2017.06.006
21. Bai S, Tian B, Li A, Yao Q, Zhang G, Li F. MicroRNA-125b promotes tumor growth and suppresses apoptosis by targeting DRAM2 in retinoblastoma. Eye (Lond). 2016;30(12):1630–1638. doi:10.1038/eye.2016.189
22. Parikh A, Lee C, Joseph P, et al. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nat Commun. 2014;5(1):2977. doi:10.1038/ncomms3977
23. Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21(5):912–916. doi:10.1038/sj.leu.2404605
24. Sun W, Wang X, Li J, et al. MicroRNA-181a promotes angiogenesis in colorectal cancer by targeting SRCIN1 to promote the SRC/VEGF signaling pathway. Cell Death Dis. 2018;9(4):438. doi:10.1038/s41419-018-0490-4
25. Wang P, Chen D, Ma H, Li Y. LncRNA SNHG12 contributes to multidrug resistance through activating the MAPK/Slug pathway by sponging miR-181a in non-small cell lung cancer. Oncotarget. 2017;8(48):84086–84101. doi:10.18632/oncotarget.20475
26. Liu Z, Sun F, Hong Y, et al. MEG2 is regulated by miR-181a-5p and functions as a tumour suppressor gene to suppress the proliferation and migration of gastric cancer cells. Mol Cancer. 2017;16(1):133. doi:10.1186/s12943-017-0695-7
27. Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–1184.
28. Lei B, Huang Y, Zhou Z, et al. Circular RNA hsa_circ_0076248 promotes oncogenesis of glioma by sponging miR-181a to modulate SIRT1 expression. J Cell Biochem. 2019;120(4):6698–6708. doi:10.1002/jcb.27966
29. Zhang L, Liu X, Che S, et al. Endometrial epithelial cell apoptosis is inhibited by a cir8073-mir181a-neurotensis pathway during embryo implantation. Mol Ther Nucleic Acids. 2019;14:262–273. doi:10.1016/j.omtn.2018.12.005
30. Hochreuter MY, Altıntaş A, Garde C, et al. Identification of two microRNA nodes as potential cooperative modulators of liver metabolism. Hepatol Res. 2019;49(12):1451–1465. doi:10.1111/hepr.13419
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.