Back to Journals » OncoTargets and Therapy » Volume 13
A Novel Circular RNA, circ_0005394, Predicts Unfavorable Prognosis and Contributes to Hepatocellular Carcinoma Progression by Regulating miR-507/E2F3 and miR-515-5p/CXCL6 Signaling Pathways
Received 31 March 2020
Accepted for publication 4 June 2020
Published 29 June 2020 Volume 2020:13 Pages 6171—6180
DOI https://doi.org/10.2147/OTT.S256238
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Chengming Sun,1 Guodong Li,2 Ming Liu2
1Department of Hepatopancreatobiliary Surgery, Cancer Hospital Affiliated to Harbin Medical University, Harbin 150001, People’s Republic of China; 2Department of General Surgery, The Fourth Affiliated Hospital of Harbin Medical University, Harbin 150001, People’s Republic of China
Correspondence: Ming Liu
Department of General Surgery, The Fourth Affiliated Hospital of Harbin Medical University, 37, Yiyuan Road, Harbin, Heilongjiang Province 150001, People’s Republic of China
Email [email protected]
Objective: Circular RNAs (circRNAs) play a key role in cancer development and progression. Previously, circ_0005394 was found to be highly expressed in hepatocellular carcinoma (HCC) screened by circRNA microarray. However, the research with regard to the functions and mechanisms of circ_0005394 in HCC remains unknown.
Materials and Methods: The expression of circ_0005394 in HCC was measured by qRT-PCR. The clinical relevance was evaluated by Fisher’s exact test, Kaplan–Meier curves, and Cox regression model. Gain/loss-of function assays were performed to elucidate the functions of circ_0005394 in Huh-7 and HepG2 cells. Dual-luciferase reporter assay was applied to reveal the mechanism of circ_0005394.
Results: circ_0005394 expression was higher in HCC tissues and cells than noncancerous samples and normal cell line, respectively. High expression of circ_0005394 was associated with larger tumor size, more advanced TNM stages, and poorer overall survival for the patients with HCC. Gain/loss-of function assays demonstrated its oncogenic role in cell growth, apoptosis, migration, and invasion. Mechanistically, miR-507 and miR-515-5p could be sponged by circ_0005394. Furthermore, E2F Transcription Factor 3 (E2F3) and C-X-C motif chemokine ligand 6 (CXCL6) were confirmed as the target of miR-507 and miR-515-5p, respectively. Rescue assay indicated that circ_0005394 facilitated HCC growth and invasion by regulating miR-507/E2F3 and miR-515-5p/CXCL6 signaling pathways.
Conclusion: This study uncovered an important role of circ_0005394 in regulating HCC progression, providing a novel perspective for clarifying its pathogenesis.
Keywords: hepatocellular carcinoma, circRNA, circ_0005394, E2F3, CXCL6
Introduction
Hepatocellular carcinoma (HCC) is a common cancer of the hepatobiliary system.1,2 Many complex factors play key roles in the development and progression of HCC cells. In recent years, dramatic advances have been made in the clinical therapy for HCC in both surgical procedure and chemotherapy regimens. However, the outcome of HCC is still unfavorable.3 Therefore, it is vital to unveil the secret of HCC carcinogenesis and metastasis, which might contribute to the discovery of new biomarkers and improve the prognosis of HCC patients.
Circular RNAs (circRNAs) are a class of endogenous non-coding RNA with covalently closed loop structures. They are not degraded by RNA exonuclease, thus maintaining a stable expression.4 They exist extensively in cells, and dysregulation of circRNAs may cause human diseases, including cancer.5 CircRNAs exert regulatory effects in multiple diseases through interacting with certain miRNAs, including esophageal cancer,6 oral cancer,7 cardiovascular disease,8,9 Alzheimer’s disease,10 etc. Such an interaction between circRNA and miRNA is known as the competitive endogenous RNA (ceRNA) theory.11 For example, circ_104718 sponges miR-218-5p to upregulate the thioredoxin domain containing 5 (TXNDC5) expression in HCC.12 Hence, determining the mechanisms that are underlying the progression of HCC and searching for new drugs from the view of circRNA is imperative. Previously, Xu et al13 identified circ_0005394 as an elevated circRNA in HCC tissues compared to the adjacent noncancerous samples. Circ_0005394 is mapped to chr16: 11868091–11876244 and generated by splicing of ZC3H7A. The spliced sequence length of circ_0005394 is 937 nucleotides long. In this work, we verified that circ_0005394 sponged miR-507 and miR-515-5p to upregulate E2F Transcription Factor 3 (E2F3) and C-X-C motif chemokine ligand 6 (CXCL6) expression levels in HCC. Moreover, miR-507/E2F3 and miR-515-5p/CXCL6 signaling pathways played a key role in mediating HCC progression.
Materials and Methods
Patients and Tissues
The HCC specimens and their adjacent normal tissues were harvested from patients who underwent liver resection at Cancer Hospital Affiliated to Harbin Medical University. A validation cohort consisting of 82 HCC patients was selected according to the following inclusion criteria: (i) patients who underwent radical resection with a clear surgical margin; (ii) patients with available follow-up information and complete medical records; (iii) patients with a survival time of more than 1 month; (iv) patients had not undergone radio/chemotherapy before surgery; and (v) patients who had no history of other malignancies. This study was authorized by the Ethics Committee of Cancer Hospital Affiliated to Harbin Medical University. Informed consent was obtained from each participant. The tumors were classified and staged according to the 8th AJCC/UICC TNM classification system. The pathological diagnosis of HCC patients was performed by two experienced pathologists. Normal tissue was collected at least 3 cm from the tumor margin. Tumor samples and its corresponding normal tissues were stored at −80°C until using.
Cell Culture and Transfection
HCC cells (Sk-Hep-1, SMMC7721, HCCLM3, Huh-7, and HepG2) and normal cell line (L02) were purchased from the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in the medium containing 10% fetal bovine serum (FBS; HyClone Laboratories, Logan, UT, USA) in a 5% CO2-humidified atmosphere at 37°C.
Small interference RNA (siRNA) target for the circ_0005394 (si-circ_0005394-1 and si-circ_0005394-2), si-E2F3, si-CXCL6, si-NC, miR-507/515-5p/NC mimics, and miR-507/515-5p/NC inhibitor were acquired from GenePharma Co., Ltd. (Shanghai, China). The overexpression vector of pcDNA-circ_0005394, E2F3, CXCL6, and pcDNA-control were brought from Hanbio Biotechnology Co., Ltd. (Shanghai, China). The targeted sequences of circ_0005394 are as follows: si-circ_0005394-1, 5′-GAAACTGTTATGATTGCTCAT-3′, and si-circ_0005394-2, 5′-AATGAAACTGTTATGATTGCT-3′. Cell transfection was conducted using Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) followed by the instructions of manufacturer. Then 125 μL serum-free medium was used to dilute 5 μL Lipofectamine 3000 in a 1.5 mL EP tube. Meanwhile, 5 μL siRNA (20 μM) or 2.5 μg plasmid vector along with 5 μL P3000™ reagent was diluted in 125 μL serum-free medium. After 5 minutes incubation at room temperature, the reagents in two tubes were combined. 15–20 minutes later, the mixtures were added into a 2.5 cm dish filled with serum-free medium. After 8 hours incubation, the medium was replaced by the medium containing 10% FBS.
qRT-PCR
Total RNA was isolated by Trizol (Thermo Fisher Scientific, Waltham, MA, USA) by the routine RNA extraction protocol. RNA concentration was detected using a Thermo Nano Drop 2000 spectrophotometer (Waltham, MA, USA). RNA quality was evaluated according to the A260/A280 ratio. RNA integrity number (RIN) analysis was performed using an Agilent 2100 Bioanalyser and RNA 6000 LabChip kit with Agilent 2100 Expert software (Agilent Technologies). The isolated RNA from tissue samples with a RIN of 7 or more was considered usable in the study. The RNA was reverse-transcribed into cDNA (Roche, Basel, Switzerland). qRT-PCR assay was performed on a 7500 fast Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) and SYBR Green Master (Roche). qRT-PCR was performed in triplicate. U6 and GAPDH were used as internal controls. Relative RNA expression was analyzed using the ΔΔCt method.
Cell Viability Detection
Transfected cells were added into 96-well plates (1500 cells per well) and maintained for indicated time. Then 10 μL of cell counting kit-8 (CCK-8) was added into each well and incubated for 2 hours. The absorbance of samples at 450 nm was detected using a microplate reader (Tecan, Männedorf, Switzerland).
Cell Apoptosis Assay
Acridine orange/ethidium bromide (AO/EB) double fluorescence staining was carried out to measure cell apoptosis. Briefly, the cells were treated with AO/EB mixed solution (Solarbio, Beijing, China). Five minutes later, the cells were counted and photographed (Leica, Wetzlar, Germany).
Annexin V‑fluorescein isothiocyanate apoptosis kit (BD Biosciences, San Jose, CA, USA) was applied to evaluate the apoptotic rate of HCC cells. In brief, the transfected cells (2 × 106) were washed with PBS twice, resuspended with 1X binding buffer, and then maintained with 5 μL of Annexin V-FITC and 10 μL of propidium iodide (PI) for 15 minutes in the dark. At last, cell apoptosis was detected using a flow cytometer (FACScan, BD Biosciences).
Wound Healing Assay
Cells were seeded into a 2.5 cm dish (2×105 cells/well). The wounds were created using a 200 μL sterile pipette tip until the cells reached 90% confluency. The cells were washed twice with PBS. At indicated culturing time, the separation distance between scratch edges was measured. The scratches were captured using a light microscope (Leica).
Transwell Assay
For cell migration detection, the bottom compartment was added with medium containing 10% FBS. The upper chamber with polycarbonate membrane (Corning, Inc., Corning, NY, USA) was filled with transfected cells in serum-free medium. After incubation, the cells were fixed by paraformaldehyde (Solarbio) and dyed by crystal violet (Beyotime, Beijing, China). The migrated cells were then imaged and counted. The protocol for the invasion experiment was similar with that in cell migration test. The difference was that the top chamber in the invasion assay was coated with Matrigel (BD Biosciences, USA).
Dual-Luciferase Reporter Gene Test
The sequences of circ_0005394 were predicted to interact with miR-507 and miR-515-5p (predicted by Circular RNA Interactome). Full length of circ_0005394 or a mutated sequence was inserted into the pmirGLO vector. Cells seeded into 24-well plates were cotransfected with luciferase vectors and miR-507/515-5p mimics or mimics-NC, and subjected to luciferase activity determination at 36 hours (Promega, Madison, WI, USA).
Data Analysis
Data are expressed as the mean ± SD and analyzed using unpaired 2-sided Student’s t-test using GraphPad Prism 6 (GraphPad, Inc., La Jolla, CA, USA). Fisher’s exact test was conducted to uncover the association between circ_0005394 expression and clinical characteristics. Kaplan–Meier curves and Cox regression model were used to measure overall survival by using SPSS 22.0 (SPSS, Inc., Chicago, IL, USA). The significance threshold was set at a P-value<0.05.
Results
circ_0005394 Is Upregulated in HCC Tissues and Cells and Correlates with Dismal Prognosis
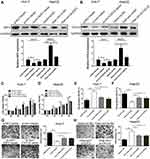
We measured circ_0005394 expression levels in HCC specimens and adjacent nontumorous samples. Upregulation of circ_0005394 was observed in HCC specimens in comparison with the normal counterparts (Figure 1A). Additionally, qRT-PCR displayed that circ_0005394 was evidently elevated in HCC cells compared to L02 cells (Figure 1B). In particular, circ_0005394 expression remained the highest in the Huh-7 cell line, but the lowest in the HepG2 cell line (Figure 1B). Afterwards, the recruited patients were divided into high/low circ_0005394 expression groups following the median value of circ_0005394. Elevation of circ_0005394 was found in HCC patients with TNM stage III–IV, relative to that in HCC samples with TNM stage I–II (P=0.007). Additionally, circ_0005394 was elevated in HCC patients with larger tumor size (P=0.013, Table 1). Furthermore, circ_0005394 expression in HCC tissue samples was markedly associated with unfavorable prognosis in HCC patients, as proved by Kaplan–Meier survival curves (P=0.002, Figure 1C). Among the clinicopathological variables, advanced TNM stages (P=0.005) and circ_0005394 expression (P=0.003) were both identified as risk factors to predict overall survival according to univariate analysis. Multivariate Cox regression analysis identified circ_0005394 as an independent prognostic predictor for HCC (P=0.039, Table 2).
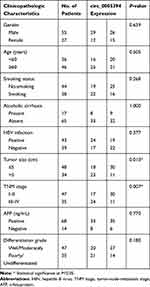 |
Table 1 circ_0005394 Expression and Clinicopathologic Characteristics of HCC Patients |
 |
Table 2 Univariate and Multivariate Analysis of Prognostic Factors for Overall Survival in HCC Patients |
circ_0005394 Promotes HCC Cell Progression
We next explored the effects of circ_0005394 on the growth, apoptosis, migration, and invasion of HCC cells. The efficiencies of siRNAs were validated and circ_0005394-siRNA-1 was selected for further study due to its high knockdown efficiency (Figure 2A). HepG2 cells was used for gain-of function assay. Figure 2B shows that circ_0005394 vector significantly enhanced circ_0005394 expression compared to the cells transfected with empty vector. For the part of functional assay, circ_0005394-siRNA mediated circ_0005394 downregulation led to lower cell viability in Huh-7 cells (Figure 2C). Elevated circ_0005394 induced higher cell viability in HepG2 cells (Figure 2D). The data of AO/EB staining and flow cytometric assays illustrated that inhibition of circ_0005394 obviously enhanced cell apoptosis (Figure 2E and G). However, circ_0005394 upregulation inhibited HepG2 cell apoptosis (Figure 2F and H). Additionally, to unveil the function of circ_0005394 in modulating the migratory and invasive capacities of HCC cells, wound scratch and transwell experiments were performed. The data unveiled that interfering circ_0005394 expression reduced the rate of migration more than the negative control cells (Figure 2I and K). On the contrary, ectopically expressed circ_0005394 could enhance HepG2 cells to migrate (Figure 2J and L). Similarly, as for the transwell invasion experiments, reducing the expression of circ_0005394 showed a decreased rate of invasion relative to the si-NC group (Figure 2K). Ectopic expression of circ_0005394 in HepG2 cells could elevate the cell invasive potential (Figure 2L).
circ_0005394 Sponges miR-507 and miR-515-5p to Upregulate E2F3 and CXCL6 Expression, Respectively
Emerging evidence has shown that circRNAs could act as a sponge to absorb miRNAs and inhibit their expression and functions in multiple malignancies.14,15 To explore the mechanisms of circ_0005394 in HCC, we constructed a competing endogenous RNA (ceRNA) model. Based on an online database of Circular RNA Interactome, miR-136, miR-153, miR-433, miR-490-5p, miR-507, and miR-515-5p were highly matched to the sequence of circ_0005394. We found that miR-507 and miR-515-5p were significantly upregulated/downregulated after circ_0005394 silencing/overexpression (Figure 3A and B). Furthermore, negative correlation of circ_0005394 and miR-507/515-5p was observed in HCC tissues (Figure 3C and D). To explore the potential interaction between circ_0005394 and miR-507/515-5p, we inserted the predicted target sites or mutated sites into the downstream of firefly luciferase gene (Figure 3E). Luciferase activity markedly decreased in HCC cells cotransfected with circ_0005394 WT and miRNA-507/515-5p mimics. However, no significant difference in luciferase intensity was seen in the cells cotransfected with circ_0005394 MUT and miRNA-507/515-5p mimics (Figure 3F–G). We also investigated the complementary miRNAs paired with 3ʹ-UTR of target mRNA by using TargetScan database. We found that upregulation of miR-507 markedly repressed E2F3 mRNA expression, while silencing of miR-507 evidently facilitated E2F3 expression (Figure 3H). Similarly, we identified that CXCL6 mRNA expression was negatively modulated by miR-515-5p (Figure 3I). Pearson’s correlation coefficient analysis illustrated the positive correlation between circ_0005394 and E2F3/CXCL6 expression in HCC tissues (Figure 3J–K). A luciferase reporter gene containing the WT or MUT E2F3 3ʹ-UTR or CXCL6 3ʹ-UTR sequences was constructed and the luciferase reporter assay was applied to confirm the predicted binding sites (Figure 3L). Following luciferase reporter gene test indicated that transfection of miR-507 mimics led to remarkable reduction of luciferase intensity of WT-E2F3 3ʹ-UTR reporter compared to that in the mimics-NC group (Figure 3M). In line with our expectation, miR-515-5p could also interact with 3ʹ-UTR of CXCL6 mRNA (Figure 3N).
circ_0005394 Executes Oncogenic Properties Through Regulating miR-507/E2F3 and miR-515-5p/CXCL6 Signalings in HCC
Rescue experiment was performed on Huh-7 and HepG2 cells to evaluate whether the malignant behaviors induced by circ_0005394 are partly attributed to its regulation of miR-507/E2F3 and miR-515-5p/CXCL6 pathways. Immunoblotting assay showed that knockdown of circ_0005394 significantly attenuated the expression levels of E2F3 and CXCL6. Whereas, after being cotransfected with E2F3 vector or CXCL6 vector, the downregulation of E2F3 and CXCL6 was partly reversed (Figure 4A and B). In addition, circ_0005394 vector cotransfected with si-E2F3 or si-CXCL6 partially reversed the elevation of E2F3 and CXCL6 caused by circ_0005394 vector (Figure 4A and B). Furthermore, CCK-8 and transwell invasion assays exhibited that the E2F3 vector or CXCL6 vector could prominently reverse the inhibition of si-circ_0005394-1 on HCC cell viability and invasion (Figure 4C and G). We found that silencing of circ_0005394 could facilitate apoptosis in Huh-7 cells, while this acceleration effect could be partly reversed by E2F3 or CXCL6 vector (Figure 4E). Furthermore, overexpression of circ_0005394 facilitated cell progression, while after cotransfection with si-E2F3 or si-CXCL6, the oncogenic properties were partially reversed in HepG2 cells (Figure 4D, F, and H).
Discussion
Accumulating studies have manifested that circRNAs act as crucial roles in multiple types of cancers, including HCC.12,15 In the previous study, the differentially expression of circRNAs was screened in HCC and normal tissues and circ_0005394 was identified as an upregulated circRNA in tumorous samples.13 In this study, circ_0005394 was found to be aberrantly elevated in HCC specimens and cells based on the data of qRT-PCR assay. For clinical value investigation, circ_0005394 expression correlated with patients’ clinical severity. In the clinical data analysis, circ_0005394 expression increased as the tumor size and TNM stage increased. Moreover, high circ_0005394 level correlated with poorer 5-year overall survival in HCC patients after surgery. According to the Cox regression model, circ_0005394 was further predicted as an independent prognostic factor for HCC patients.
Functional experiments indicated that downregulation of circ_0005394 inhibited HCC cell growth by triggering cell apoptosis. Furthermore, decreased circ_0005394 repressed HCC cell migration and invasion. By contrast, circ_0005394 overexpression induced the opposite effect to that mentioned above. After validation of the clinical value and functional role of circ_0005394, we investigated the mechanism of circ_0005394 in HCC. CeRNA hypothesis proposes a novel mechanism in the interaction of RNAs.16,18 Recent studies identified the regulatory functions of circRNAs in tumor cell behaviors as ceRNAs.19,20 Here, we predicted several miRNAs which may interact with circ_0005394 through the online database. Furthermore, qRT-PCR and dual-luciferase reporter assay indicated that only miR-507 and miR-515-5p were sponged and modulated by circ_0005394. These two miRNAs were reported as tumor suppressors in previous studies.21,26 For miR-507, it suppresses breast cancer cell aggressiveness by targeting fms related receptor tyrosine kinase 1 (Flt-1).21 Additionally, another report indicated that miR-507 is regulated by a lncRNA, urothelial cancer associated 1 (UCA1), thereby regulating forkhead box M1 (FOXM1) expression at posttranscriptional level.22 For miR-515-5p, it suppresses prostate cancer cell progression via targeting thyroid hormone receptor interactor 13 (TRIP13).23 In non-small cell lung cancer (NSCLC), CXCL6 could promote cell survival and metastasis through downregulating miR-515-5p.24 Furthermore, a higher level of miR-515-5p associates with increased breast cancer patients’ survival.25 In gastric cancer, miR-515-5p/X-Linked inhibitor of apoptosis (XIAP) signaling is modulated by hsa_circ_0067997.26 In this project, the results revealed that circ_0005394 acted as a sponge to absorb miR-507 and miR-515-5p. We next identified E2F3 and CXCL6 as the direct target of miR-507 and miR-515-5p, respectively. E2F3 belongs to the E2F family and binds to DP interaction partner proteins to exert its functions.27 It is imperative for E2F3 to modulate cellular growth of both primary and tumor cells via induction of apoptosis by DNA damage.28,29 What’s more, E2F3 exerts oncogenic function in various cancers, including HCC.30 CXCL6, also known as granulocyte chemotactic protein 2, was firstly found in MG63 cells, one of the osteosarcoma cell lines. CXCL6 regulates its downstream pathways via binding withC-X-C Motif Chemokine Receptor 1 (CXCR1) and C-X-C Motif Chemokine Receptor 2 (CXCR2).31 Growing evidence has demonstrated that CXCL6 plays a key role in mediating tumorigenesis. For instance, theactivated CXCL6/CXCR1/2 pathway facilitates the growth and metastasis of osteosarcoma cells.32 In the current study, we for the first time revealed that the oncogenic function of circ_0005394 is partly dependent on its upregulation of E2F3 and CXCL6, which broadened the perspective of circRNAs in HCC.
However, there are still some limitations within the study. Animal study was not performed and a larger cohort of patients and multicenter study are essential to validate the clinical significance of circ_0005394 in the future studies.
Conclusions
To sum up, this study proved that circ_0005394 functioned as a ceRNA to facilitate HCC progression via sponging miR-507 and miR-515-5p, thereby elevating E2F3 and CXCL6 expression, respectively. We propose that circ_0005394 may play an important role in HCC growth and metastasis.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477–491. doi:10.1053/j.gastro.2018.08.065
2. Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin Gastroenterol Hepatol. 2015;13(12):2140–2151. doi:10.1016/j.cgh.2015.08.014
3. Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. doi:10.3322/caac.21349
4. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
5. Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496(2):455–461. doi:10.1016/j.bbrc.2018.01.077
6. Su H, Lin F, Deng X, et al. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14(1):225. doi:10.1186/s12967-016-0977-7
7. Chen L, Zhang S, Wu J, et al. circRNA_100290 plays a role in oral cancer by functioning as a sponge of the miR-29 family. Oncogene. 2017;36:4551–4561.
8. Tan WL, Lim BT, Anene-Nzelu CG, et al. A landscape of circular RNA expression in the human heart. Cardiovasc Res. 2017;113(3):298–309. doi:10.1093/cvr/cvw250
9. Zou M, Huang C, Li X, et al. Circular RNA expression profile and potential function of hsa_circRNA_101238 in human thoracic aortic dissection. Oncotarget. 2017;8(47):81825–81837. doi:10.18632/oncotarget.18998
10. Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is linked to deficits in a natural circular miRNA-7 sponge (circRNA; ciRS-7). Genes (Basel). 2016;7.
11. Li LJ, Zhao W, Tao SS, et al. Competitive endogenous RNA network: potential implication for systemic lupus erythematosus. Expert Opin Ther Targets. 2017;21:639–648. doi:10.1080/14728222.2017.1319938
12. Yu J, Yang M, Zhou B, et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond). 2019;11:8182–8203.
13. Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):98. doi:10.1186/s13046-019-1041-2
14. Mao W, Huang X, Wang L, et al. Circular RNA hsa_circ_0068871 regulates FGFR3 expression and activates STAT3 by targeting miR-181a-5p to promote bladder cancer progression. J Exp Clin Cancer Res. 2019;38(1):169. doi:10.1186/s13046-019-1136-9
15. Li X, He M, Guo J, Cao T. Upregulation of circular RNA circ-ERBB2 predicts unfavorable prognosis and facilitates the progression of gastric cancer via miR-503/CACUL1 and miR-637/MMP-19 signaling. Biochem Biophys Res Commun. 2019;511(4):926–930. doi:10.1016/j.bbrc.2019.03.010
16. Bossi L, Figueroa-Bossi N. Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria. Nat Rev Microbiol. 2016;14(12):775–784. doi:10.1038/nrmicro.2016.129
17. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–10100. doi:10.1002/jcp.27941
18. Zhu Y, Bian Y, Zhang Q, et al. Construction and analysis of dysregulated lncRNA-associated ceRNA network in colorectal cancer. J Cell Biochem. 2019;120(6):9250–9263. doi:10.1002/jcb.28201
19. Zhang Y, Ke X, Liu J, et al. Characterization of circRNA-associated ceRNA networks in patients with nonvalvular persistent atrial fibrillation. Mol Med Rep. 2019;19(1):638–650. doi:10.3892/mmr.2018.9695
20. Wang WL, Yang Z, Zhang YJ, et al. Competing endogenous RNA analysis reveals the regulatory potency of circRNA_036186 in HNSCC. Int J Oncol. 2018;53(4):1529–1543. doi:10.3892/ijo.2018.4499
21. Jia L, Liu W, Cao B, Li H, Yin C. MiR-507 inhibits the migration and invasion of human breastcancer cells through Flt-1 suppression. Oncotarget. 2016;7(24):36743–36754. doi:10.18632/oncotarget.9163
22. Wei Y, Sun Q, Zhao L, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016;33(8):88. doi:10.1007/s12032-016-0804-2
23. Zhang X, Zhou J, Xue D, Li Z, Liu Y, Dong L. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int J Biol Macromol. 2019;129:227–232. doi:10.1016/j.ijbiomac.2019.01.127
24. Li J, Tang Z, Wang H, et al. CXCL6 promotes non-small cell lung cancer cell survival and metastasis via down-regulation of miR-515-5p. Biomed Pharmacother. 2018;97:1182–1188. doi:10.1016/j.biopha.2017.11.004
25. Pardo OE, Castellano L, Munro CE, et al. miR-515-5p controls cancer cell migration through MARK 4 regulation. EMBO Rep. 2016;17(4):570–584. doi:10.15252/embr.201540970
26. Zhang H, Wang X, Huang H, Wang Y, Zhang F, Wang S. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR-515-5p and activation of X chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells Nanomed Biotechnol. 2019;47(1):308–318. doi:10.1080/21691401.2018.1553787
27. DeGregori J. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochim Biophys Acta. 2002;1602(2):131–150. doi:10.1016/s0304-419x(02)00051-3
28. Martinez LA, Goluszko E, Chen HZ, et al. E2F3 is a mediator of DNA damage-induced apoptosis. Mol Cell Biol. 2010;30(2):524–536. doi:10.1128/MCB.00938-09
29. Nahle Z, Polakoff J, Davuluri RV, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4(11):859–864. doi:10.1038/ncb868
30. Han R, Chen X, Li Y, Zhang S, Li R, Lu L. MicroRNA-34a suppresses aggressiveness of hepatocellular carcinoma by modulating E2F1, E2F3, and Caspase-3. Cancer Manag Res. 2019;11:2963–2976. doi:10.2147/CMAR.S202664
31. Wolf M, Delgado MB, Jones SA, Dewald B, Clark-Lewis I, Baggiolini M. Granulocyte chemotactic protein 2 acts via both Il-8 receptors, Cxcr1 and Cxcr2. Eur J Immunol. 1998;28(1):164–170. doi:10.1002/(SICI)1521-4141(199801)28:01<164::AID-IMMU164>3.0.CO;2-S
32. Liu G, An L, Zhang H, Du P, Sheng Y. Activation of CXCL6/CXCR1/2 axis promotes the growth and metastasis of osteosarcoma cells in vitro and in vivo. Front Pharmacol. 2019;28:307. doi:10.3389/fphar.2019.00307
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.