Back to Journals » Drug Design, Development and Therapy » Volume 17
A Novel Approach Based on Gut Microbiota Analysis and Network Pharmacology to Explain the Mechanisms of Action of Cichorium intybus L. Formula in the Improvement of Hyperuricemic Nephropathy in Rats
Authors Amatjan M, Li N, He P, Zhang B, Mai X, Jiang Q, Xie H , Shao X
Received 30 September 2022
Accepted for publication 11 January 2023
Published 20 January 2023 Volume 2023:17 Pages 107—128
DOI https://doi.org/10.2147/DDDT.S389811
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Tin Wui Wong
Mukaram Amatjan,1 Na Li,1 Pengke He,1 Boheng Zhang,1 Xianyan Mai,1 Qianle Jiang,1 Haochen Xie,2 Xiaoni Shao1
1Immunotherapy Laboratory, College of Pharmacology, Southwest Minzu University, Chengdu, 610225, People’s Republic of China; 2Qinghai Tibet Plateau Research Institute, Southwest Minzu University, Chengdu, 610225, People’s Republic of China
Correspondence: Xiaoni Shao, Immunotherapy Laboratory, College of Pharmacology, Southwest Minzu University, #168 Dajian Street, Shuangliu District, Chengdu, 610225, People’s Republic of China, Tel +86-28-85738423, Email [email protected]
Background: Cichorium intybus L. formula (CILF) is a traditional Chinese medicine (TCM) widely used in the treatment of gout and hyperuricemic nephropathy (HN). The aim of this research was to investigate the potential protective effect of CILF against HN and elucidated the underlying mechanism.
Methods: CILF water extract was administered to an HN rat model established by adenine combined with ethambutol. The levels of uric acid (UA), serum urea nitrogen (UREA), and creatinine (CREA) were detected. Changes in the pathology and histology of the kidney were observed by hematoxylin-eosin staining. The 16S rRNA of the gut microbiota was sequenced. The binding ability of the main ingredients of CILF to key targets was analyzed by network pharmacology and molecular docking. The expression levels of the related mRNAs and proteins in the kidney were evaluated by RT-qPCR and immunohistochemistry analysis.
Results: CILF administration significantly alleviated increases in UA, UREA, and CREA, structural damage, and kidney dysfunction. Gut microbiota analysis was applied to explore the pharmacological mechanism of the effects of CILF on bacterial diversity and microbiota structure in HN. CILF decreased the abundance of Bacteroides. In addition, it increased the abundance of Lactobacillaceae, Erysipelotrichaceae, Lachnospiraceae, Ruminococcaceae, and Bifidobacterium. Based on network pharmacology and molecular docking analysis, CILF profoundly influenced the IL17, TNF and AGE-RAGE signaling pathway. Additionally, CILF inhibited the expression of STAT3, VEGFA and SIRT1 to improve the symptoms of nephropathy. Our research suggested that CILF protects against kidney dysfunction in rats with HN induced by adenine combined with ethambutol.
Conclusion: Our findings on the anti-HN effects of CILF and its mechanism of action, from the viewpoint of systems biology, and elaborated that CILF can alter the diversity and community structure of the gut microbiota in HN, providing new approaches for the prevention and treatment of HN.
Keywords: Cichorium intybus L. formula, hyperuricemic nephropathy, gut microbiota, network pharmacology, molecular docking
Introduction
Hyperuricemia can lead to chronic kidney disease (CKD) and end-stage renal disease. The main pathological basis is that luminescent crystals are deposited in the kidney collecting tubules, resulting in acute renal injury and renal disease.1 Due to the complex pathogenesis of hyperuricemic nephropathy (HN), the lack of any effective clinical treatment will allow the disease to progress. Adenine increased serum UREA and CREA concentrations and excretion of UA in urine, produced proteinuria and induced kidney atrophy and fibrosis.2 At the same time, ethambutol can inhibit the secretion of UA by the renal tubules, thereby inhibiting the excretion of UA by the kidneys and increasing serum UA, leading to kidney damage.3 Previous reports have suggested that adenine in combination with ethambutol induces HN.4,5 Renal damage caused by inducing chronic high levels of UA is sufficient to cause HN. Currently, the treatment of HN is mainly achieved by reducing UA levels and improving the symptoms of kidney disease.6
There are a number of Western medicines available for the treatment of HN, such as febuxostat, allopurinol and benzbromarone.7–9 Although they are very effective, they inevitably produce varying degrees of adverse effects, such as fatal hepatic failure and hypersensitivity syndrome.10,11 Compared to Western medicines, traditional Chinese medicine (TCM) has the advantages of milder action and fewer adverse effects.12 TCM is an unmet medical need in HN clinical research. According to TCM theory, defects in the spleen and kidney are the fundamental pathogenesis of HN, resulting in the failure of grain and water transportation and transformation, as well as the blockage of dampness and phlegm in meridian.13 Therefore, clinical treatment focuses on dampness-heat, blood stasis and deficiency of the spleen and kidney. Resolving phlegm and turbidity, promoting blood circulation and removing blood stasis, tonifying the kidney and supplementing Qi are the basic treatment principles of hyperuricemic renal injury. HN is accompanied by severe renal function injury and reduced UA excretion.14 Cichorium intybus L. formula (CILF) consists of seven herbs, chicory (Cichorium intybus L.), gardenia (Gardenia jasminoides Ellis), radix puerariae (Pueraria lobata (Willd.) Ohwi), lilium (Lilium brownii F.E. Brown var. viridulum Baker), mulberry leaf (Morus alba L.), angelica dahurica (Angelica Dahurica (Fisch, Ex Hoffm. Benth. et hook.f.), and dandelion (Taraxacum officinale Weber).
Chicory and gardenia have anti-hyperglycemic, anti-inflammatory, antioxidant, and significant effects on the digestive system.15,16 Radix puerariae, lilium and angelica dahurica have antioxidant, anti-inflammatory and antibacterial effects.17–19 Mulberry leaves and dandelion have protective effects against kidney damage, and their positive effects on the gut microbiota have been reported.20,21 In summary, the seven TCM components regulate metabolism, protect kidney function and inhibit inflammation. At the same time, the abovementioned formula is both food and TCM, meaning it can be used more effectively. Therefore, this study speculated that the combination of the seven drugs would have a better effect on reducing UA levels, kidney injury and inflammation, but there is no relevant report on the combination of the seven drugs mentioned above.
The relationship between gut microbiota and metabolic diseases, especially HN, has also become a research hotspot. It has been reported that some products produced by gut microbiota metabolism can affect the level of UA. There is mounting evidence that the pathophysiological status of the host and microenvironment is correlated with the microbiome in the gut of CKD patients.22 The “intestine-kidney-axis” theory explains that the interactions between the gut microbiota and CKD are bidirectional.23 The impairment of renal function leads to disorders of the gut microbiota. It can change the gut microbiota by destroying the intestinal mucosal barrier and transferring endotoxin and bacterial metabolites into the systemic circulation through the intestinal barrier, inducing chronic inflammation and accelerating kidney damage. This shows that under this situation, the intestine, which is responsible for the excretion of 30% of UA, will also be an important target organ for the development of new drugs to improve HN.
TCM has the characteristics of numerous components and complex mechanisms, and it is difficult to reflect the systematization of Chinese medicine by studying single components and targets. In recent years, the research method of network pharmacology on multicomponent, multitarget and multichannel TCM formulas has gradually attracted attention. Through the analysis of network pharmacology and molecular docking, the binding activity between the potential pharmacodynamic components of the compound and the key targets is screened, and the chemical substances and action mechanism closely related to the functional attributes of its treatment are found in the formula.24
Therefore, this study investigated the effects of CILF on adenine combined with ethambutol-induced HN in rats and its potential mechanisms. Building on previous research,25 our group added two herbs to improve efficacy. The values of UA, CREA and UREA were significantly improved by CILF treatment. RT-PCR was validated using different targets, and immunohistochemistry was used to verify the possible mechanism of CILF in hyperuricemia renal impairment. At the same time, 16S rRNA sequencing analysis was used to study the effect of CILF on intestinal flora, and a significant increase in the abundance of microflora was found. It is proved that CILF is a seven-flavored herb with better effect, which can reduce the absorption of various inflammatory substances and bidirectional synergistic improvement in HN around the “intestine-kidney-axis”.
Materials and Methods
Preparation of CILF Water Extract
All constituent herbs in CILF in Table 1 were obtained from Madison Co., Ltd. (Xinjiang, China) and were identified by Professor Jiachuan Li. Voucher specimens are kept at the herbarium of the College of Pharmacy, Southwest Minzu University (No. 20228110190J). Preparation dried herbs of CILF (58 g) were decocted twice with a ten-fold mass of water to boil for 2 h. After filtration, the filtrates were combined and then concentrated using a rotating evaporator to configure it to the required concentration, and lyophilized in a freeze-dryer. The CILF extract was stored at 4°C until use; it was then dissolved in distilled water for use.
 |
Table 1 Medicinal Plants Contained in CILF Decoction |
UHPLC-MS Analysis
The main chemical constituents of CILF were characterized by UPLC-Q/Orbitrap HRMS, and LC separation was performed on a Hypersll GOLD C18 column (100 × 2.1 mm, 3 μm, Thermo Fisher Scientific, CA, USA). The column temperature was maintained at 30°C. The mobile phase was composed of 0.1% acetonitrile (A) and 0.1% formic acid/water (B) at a flow rate of 0.3 mL/min. The gradient elution program was as follows: 0~2 min:2% A→5% A;2~5 min:5% A→10% A;5~12 min: 10% A→16% A. The detection wavelength was set at 254 nm.
Mass spectrometry analysis was performed on a Q-Exactive Plus™ mass spectrometer (Thermo Fisher). The MS instrument was connected to UHPLC via a heated electrospray ionization (HESI) interface. The HESI source was operated in ion mode. The carrier was nitrogen, while the sheath gas and auxiliary gas pressures were MPa and 1.0 MPa, respectively. The spray voltage was 3.80 kV in positive mode and 3.00 kV in negative mode. The capillary temperature was set to 350 °C and the auxiliary heater to 200 °C. The resolution was 70,000 for full MS and 17,500 for data-dependent MS2. Compounds were detected at a mass-to-charge-ratio of 100 to 1500 m/z, and fixed collision energies were used (20, 30, 40 eV). Data were processed using Xcalibur™ 4.1 software (Thermo Fisher).
Chemicals and Reagents
Ethambutol was purchased from Chengdu Jinhua Pharmaceutical Co., Ltd. (Sichuan, China). Adenine was purchased from the Aladdin company (Shanghai, China). Allopurinol tablets are obtained from HeJulianlian Pharmaceutical Co., Ltd. (Anhui, China). Formic acid (LC-MS grade) and acetonitrile were acquired from Tedia Company (Ohio State, USA).
Animals and Drug Administration
Thirty male Wistar rats, (200 ± 10g, 7–8 weeks) were provided by Chengdu Dossy Experimental Animal Co., Ltd. (Sichuan, China), No. [SCXK (Sichuan) 2015–030], and housed in the SPF Animal Laboratory of Southwest Minzu University (Sichuan, China) at a temperature of 23 ± 2°C and relative humidity of 50 ± 10% with a 12 h/12 h light/dark cycle. During the study, the rats were given free access to food and water. The animal protocol used in this study has been reviewed and approved by the animal ethics committee of Southwest Minzu University (Approval No. 2022–09). All experiments were treated by the Regulations of Animal Administration issued by the State Committee of Science and Technology of the People’s Republic of China.
Rats were orally administrated by combined adenine and ethambutol to induce HN, as described previously.26 The HN model was administered intragastrically by combining adenine (100 mg/kg) and ethambutol (250 mg/kg) once daily for 21 days. All 30 male rats were divided into five groups: control group (CG), HN group (HNG), allopurinol group (AG, 50 mg/kg/day), high-dose CILF treatment group (CHG, 10.44 g/kg/day) and low-dose CILF treatment group (CLG, 2.61 g/kg/day) for intragastric administration. One week after induction of HN, 0.9% normal saline, allopurinol and CILF were given continuously every 24 hours.
After 3 weeks of administration, the intestinal feces were collected under sterile conditions and then preserved at −80°C. All rats fasted for 12 h, rats were anesthetized with 3% pentobarbital sodium and then whole blood was withdrawn. Kidney tissue samples were harvested. The kidney tissues were fixed with 4% paraformaldehyde and stored at 4°C. Blood samples were harvested for further analysis.
Serum Biochemistry Analysis
Blood samples were obtained by abdominal vein and centrifuged at 3000 rpm for 15 min to obtain serum. Clinical chemistry analysis of the serum was carried out on a COBAS C311 automatic biochemistry analyzer (Roche, Switzerland) using appropriate kits with the following parameters: UA, CREA, and UREA.
Histological Evaluation
The kidney tissues were fixed with 10% paraformaldehyde were dehydrated through an alcohol concentration gradient and were then cleared in xylene, embedded in paraffin, and finally sliced into 4 µm thick sections. To observe structural damage to renal tissue, sections were stained with hematoxylin and eosin (H&E), The sections were visualized using an upright optical microscope (Eclipse E100, Nikon Instruments Inc., Japan), and stained slides were captured and analyzed.
Microbial 16S rRNA Gene Sequence and Metagenomic Analyses
Fresh fecal samples were collected for 16S rRNA gene sequencing. The total microbial DNA was extracted by using a BIOMICS DNA Microprep Kit (Zymo Research, California, USA) by the manufacturer’s instructions. The 16S rDNA V4 region was amplified with the primers 515F (5’-GTGYCAGCMGCCGCGGTAA-3’) and 806R (5“-GGACTACHVGGGTWTCTAAT-3”) by a thermocycler PCR system (GeneAmp 9700, ABI, USA). The resulting RT-qPCR products were extracted and purified using the Zymoclean Gel Recovery Kit (Zymo Research, California, USA) and were quantified using Qubit@ 2.0 fluorometer (Thermo Fisher Scientific, USA) according to the manufacturer’s protocol. The purified amplicons were sequenced (PE250) using an Illumina HiSeq Rapid SBS Kit v2 (Illumina, CA, USA).
Based on Usearch software (http://drive.com/uparse/), the UPARSE algorithm was used to conduct OTU clustering at 97% consistency level, and selects the sequence with the highest frequency in each OTU as the representative sequence of OUT. Use R language (v3.6.0) for various data conversions and the ggplot2 package is used for drawing. Alpha diversity and beta diversity analyses were performed using the R language. For alpha diversity analysis, including the Chao1, PD, Shannon index, and Simpson index were calculated. For beta diversity analysis, principal coordinate analysis (PCoA) and Non-metric Multi-Dimensional Scaling (NMDS) were performed. Use the Lefse tool (https://bitbucket.org/biobackery/biobakery/wiki/Home) to find important species that differ significantly between groups. Random forest analysis uses the R language of the Random Forest database. Metastasis analysis uses R-language scripts. The differential abundance of gut microbiota between rat groups was tested by linear discriminant analysis (LDA) coupled with effect size measurement (LefSe) analysis and metagenomeSeq analysis. The sequencing data of the 16S rRNA gene were clustered and annotated based on the SILVA database, and the results are linearly converted to the classification spectrum based on KEGG database microorganisms based on the pre-calculated correlation matrix.
Network Pharmacology Profiling
To construct chemical ingredients of the CILF database, according to the Latin names of seven components, the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) (http://ibts.hkbu.edu.hk/LSP/tcmsp.php) was collected information about the chemical composition, both oral bioavailability (OB) ≥30% and drug-likeness (DL) ≥0.18 to obtain potential active components. In addition, the SMILES format structure information of compounds was downloaded from PubChem (https://pubchem.ncbi.nlm.nih.gov/). Swiss Targets Prediction (http://www.swisstargetprediction.ch/) were employed to collect ingredient-related targets by searching their computed descriptors or uploading SMILES structures.27 The known therapeutic targets acting on HN were searched and collected from databases including the DrugBank database (http://www.drugbank.ca/), Disgenet (http://www.disgenet.org/), GeneCards (https://www.genecards.org/) and Online Mendelian Inheritance in Man (OMIM, http://www.omim.org/).28 An integrative analysis of CILF, directly related genes, and HN targets was performed using a Venn analysis.
To analyze the relationship between ingredients and these potential therapeutic targets, a network was constructed and visualized among herbs, active ingredients and putative targets of CILF using Cytoscape software (version 3.8.0, http://www.cytoscape.org/).29 The protein-protein interaction (PPI) network of potential targets was constructed with the STRING (https://string-db.org/) database. The species was limited to “Homo sapiens” and the confidence score was set at 0.400. Next, predicted direct and functional target genes from the STRING data were collected to establish a PPI network. The results of PPI network construction were imported into Cytoscape software and identified with the core targets in the PPI network were identified.28
Gene Ontology (GO) functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed by loading the clusterProfiler package in R language for prospective targets of active components. Threshold p ≤ 0.05 statistics were screened for biological processes and pathways with significant differences and bar graphs were performed.30
Molecular Docking
The 3D structures of ligands were obtained from PubChem, the energy was minimized in ChemOffice software and the structure was saved in mol2 format. The crystal structures of receptors were downloaded from RCSB Protein Data Bank (https://www.rcsb.org/). PyMOL software was used to affix hydrogen atoms and remove water molecules from proteins, and the format of receptors and ligands was transformed into PDBQT through AutoDockTools 1.5.6 software. Docking simulations were performed with selected targets using the AutoDock Vina platform, the conformation with the best binding affinity was selected as the final docking conformation and visualized by PyMOL software.31
Quantitative Real-Time PCR
Total RNA was extracted from the frozen kidney samples using the Animal total RNA isolation kit (Sichuan Fujian, China) according to the manufacturer’s instructions. cDNA was generated with 2 μg of total RNA using a 5×All-In-One MasterMix with AccuRT Genomic DNA Removal kit (ABM, China). RT-qPCR was performed on an EvaGreen Express 2×qPCR with MasterMix-No Dye (ABM, China) in a 20 μL reaction volume. RT-qPCR was performed using primers specific to the target genes (Table S1). The cycling program was set as follows: the initial cycle of 95°C for 10 min, followed by 40 cycles of 95°C for 10s and 60°C for 30s. Relative gene expression was calculated by using the ∆∆CT method using the equation 2−∆∆Ct. β-Actin was amplified in a parallel reaction as an internal quantitative control.
Immunohistochemistry Analysis
The paraffin sections were deparaffinized; washed sequentially with xylene, 85% alcohol, 75% alcohol, and distilled water; then used for antigen retrieval with citric acid antigen retrieval buffer, and were then incubated with 3% H2O2 for 10 min to quench endogenous peroxidase activity. Subsequently, blocked with goat serum albumin at room temperature for 20 minutes. The sections were then incubated overnight at 4°C with diluted primary antibodies against SIRT1, STAT3 (A17307, A5511, ABclonal, Wuhan, China) and VEGFA (AF6204, Affinity, Jiangsu, China) 1:100 dilution respectively. After washing 3 times with phosphate-buffered solution (PBS), and dropped addition the secondary antibody. Then, the slices were washed 3 times with PBS, and a freshly prepared diaminobenzidine (DAB) chromogenic solution was added, dehydrated, and fixed. The tissue sample slides obtained from different groups were observed under a digital biological microscope (BA-400, MOTIC, Fujian, China). Images were analyzed using Case Viewer 2.4 software, and brown staining values were expressed as the positive staining rates.32
Statistical Methods
First, the normality of data is tested by Shapiro–Wilk test together with QQ plot. For normally distributed data, a one-way analysis of variance (ANOVA) followed by a Tukey post hoc test was used. For non-normally distributed data, the Kruskal–Wallis test was used. Data conforming to a normal distribution are expressed as mean ± SD, and skewed data are presented as median (min-max). GraphPad Prism 8.0 software (GraphPad, CA, USA) and SPSS 24.0 were used for statistical analysis and image construction. p < 0.05 was considered statistically significant.
Results
Chemical Composition of CILF
To identify the main compounds in CILF, a total of 12 compounds were characterized in CILF by UPLC-Q/Orbitrap HRMS (Figure 1 and Table S2). In our analysis, 12 compounds, neochlorogenic acid, chlorogenic acid, cryptochlorogenic acid, isochlorogenic acid B, isochlorogenic acid A, and isochlorogenic acid C in chicory; shanzhiside and genipin 1-gentiobioside in gardenia; and puerarin, puerarin 6”-O-xyloside and daidzin ether in puerariae lobatae radix showed peaks in the liquid chromatogram.
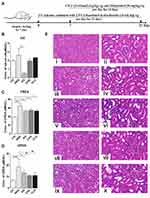
CILF Alleviates Hyperuricemic and Renal Impairment in Rats
HN model established by intragastric administration of adenine combined with ethambutol for 21 days (Figure 2A). As shown in Figure 2B–D, UA, CREA and UREA levels in HNG serum were significantly increased. The high and low doses of CILF restored these indices. UA accumulation in the kidney can lead to chronic inflammation and further renal injury. Clinically, CREA and UREA are regarded as indicators of kidney injury.33,34
Effects of CILF on Histopathologic Changes in Kidney Tissues in HN Rats
As shown in Figure 2E, the morphology and structure of the renal tissue capsule of CG rats were complete; no connective tissue hyperplasia or inflammatory exudation was observed; there was no degeneration, necrosis or fibrosis of the glomerulus in the cortical area; and no inflammatory cell infiltration was observed in the interstitium (Figure 2Ei and ii). The HNG kidney capsule was intact, the glomerular structure was normal, and the basement membrane was thickened. There was considerable fibrous tissue proliferation in the interstitium of the lesion area, and the fibroblast nuclei were fusiform with considerable inflammatory cell infiltration (Figure 2Eiii and iv). AG, CHG and CLG can significantly improve the thickening of the basement membrane, local interstitial cell infiltration, and fibrous tissue hyperplasia in the lesion area and reduce the pathological degree of the lesion (Figure 2Ev–x), suggesting that CILF has a certain effect on improving kidney injury and protecting renal function.
Gut Microbiota Analysis
In this study, CILF improved the symptoms of adenine combined with ethambutol-induced HN in rats. The intestine, which is responsible for 1/3 of UA excretion, is an important target organ for the development of new UA-lowering drugs. Therefore, gut microbiota analysis after CILF treatment was performed. First, in order to obtain a high-quality sequence, it is necessary to conduct quality control on the sequences, and the resulting sequences after quality control are called clean tags. The results are assessed by calculating the original PE, original tag, clean tag, effective tag and effective rate (%) for each sample using the mean length (NT) and Q30 (%).
To further observe the effect of CILF on the gut microbiota of rats with HN, we investigated the microbial species and their relative abundance at the phylum, class, order, family and genus classification levels according to the operational taxonomic units (OTUs). As shown in Figure 3A–C, the community structure and abundance of all bacteria in CG, HNG, Ag, CHG and CLG were very similar, mainly comprised of Firmicutes and Bacteroidetes. Compared to the CG, the number of Burkholderiaceae and Erysipelotrichaceae increased, Ruminococcaceae and Prevotellaceae decreased in the HNG. The composition and structure of the gut microbiota of rats in the CHG and CLG groups were significantly changed compared with those in the HNG group. Lactobacillaceae and Erysipelotrichaceae relative to HNG decreased, CHG and CLG treatment tended to gradually restore these indices, and the abundance of Lachnospiraceae and Burkholderiaceae in HNG increased. At the genus level, Ruminococcaceae UCG-005 and Ruminococcaceae UCG-014 of HNG showed a downward trend compared to CG. The number of Lactobacillus was found to be relatively high in CHG and CLG, while the number of Catenibacterium and Bifidobacterium also showed an upward trend compared with CG, HNG, and AG. The results showed that CILF could increase the number of beneficial bacteria, such as intestinal Lactobacillus.
To display the data distribution characteristics of the group samples, a box diagram was used for community composition analysis.35 The genus with the highest abundance was selected to display in the boxplot (Figure 3D and E). Heatmaps were used for community composition analysis to show more species groups and to compare their variation from sample to sample (Supplementary Figure S1).
To determine whether species were common or unique among groups or between samples, a Venn plot was used for community analysis. An OTU abundance table was used to make a Venn diagram, and each set was counted according to the existence of the OTUs among each sample group. In our study, there were 992 shared OTUs among the 5 groups and there were 611, 235, 538, 246, and 171 unique OTUs in the CG, HNG, AG, CHG, and CLG, respectively (Supplementary Figure S2A). To easily display the classification level information, the species abundance information and the comparisons between samples, a specific species classification tree was developed (Supplementary Figure S2B). As shown in Supplementary Figure S2C and D, to show which species have greater fluctuations in abundance, the coefficient of variation (CV) was used to measure the degree of variation in species abundance.36 To show the distribution of each species in the sample, the OTU frequency analysis was used to observe the abundance of the species present in different samples.37
The richness, evenness and diversity of microbial communities can be reflected by calculating the alpha diversity index of the samples. The observed species index and Chao1 index were used to calculate the abundance of species; the Shannon index, Simpson index, and PD whole tree index were used to calculate the diversity of species. The α diversity index of the gut microbiota is shown in Figure 4A–D. Chao1, Shannon, Simpson and PD indices all had higher CG than HNG, CHG and CLG were lower than HNG, and PD values had lower CLG than the other groups. Moreover, these index results can be further verified by the results of the rarefaction curve (Figure 4E) and rank abundance curve (Figure 4F). Therefore, CILF has a significant inhibitory effect on bacterial diversity.
To conveniently observe the different degree and variation law between samples, principal coordinates analysis (PCoA) was performed with beta diversity analysis. The PCoA results showed that the points of the CHG and CLG were widely distributed between the CG and HNG, which means that CILF can actively recover the structure of the gut microbiota to that observed in the CG. However, AG did not show a consistent positive regulatory effect compared with HNG (Figure 5A–D). The NMDS analysis based on β diversity reflects differences in the distance between samples’ passing points. The smaller the stress value is, the better. The CG, HNG, AG, CHG and GLG samples did not overlap (Figure 5E–H), indicating that there were differences among the gut microbiota components in each group.
We performed LEfSe analysis to explore the important species with significant differences in the key microbiota. Biomarkers were certified using the LDA score of Lefse and the cladogram of Lefse, and LDA values exceeding 3.5 were considered the screening criteria for major microorganisms. Ruminococcaceae UCG-005 and Ruminococcaceae UCG-014 are the primary microorganisms of CG; Erysipelotrichaceae, Actinobacteria, Bifidobacteriaceae and Catenibacterium are the main microorganisms of CHG; and AG has only one dominant microorganism, namely, Bacteria (Figure 6A and B). These results revealed that CILF treatment significantly affected the gut microbiota.
There were significant differences between the CG and HNG with regard to the microbiota structures, specifically for: Actinobacteria, Tenericutes, Spirochaetes and Epsilonbacteraeota at the phylum level; Lactobacillaceae, Burkholderiaceae, Erysipelotrichaceae, Ruminococcaceae, Prevotellaceae, Bifidobacteriaceae, and Bacteroidaceae at the family level; and Lactobacillus, Ralstonia, Lachnospiraceae NK4A136 group, Catenibacterium, Bifidobacterium, Ruminococcaceae UCG-014, Prevotellaceae Ga6A1 group, Bacteroides, Lachnospiraceae UCG-008, and Ruminococcaceae UCG-005 at the genus level.
After CILF administration, the following significant differences were identified between HNG and the CILF treatment groups for the microbiota structure: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria at the phylum level; Lactobacillaceae, Burkholderiaceae, Lachnospiraceae, Erysipelotrichaceae, and Bifidobacteriaceae at the family level, Lactobacillus, Ralstonia, Lachnospiraceae NK4A136 group, Catenibacterium, Bifidobacterium, Ruminococcaceae UCG-014, Prevotellaceae Ga6A1 group, Bacteroides, and Lachnospiraceae UCG-008 at the genus level. According to the results, these changes in colony abundance are closely related to the structural changes in intestinal flora in CILF rats before and after administration.
Random forest analysis and different tests can be combined to find species with significant differences between groups. It can also be used to find important biomarkers. There were 35 species of flora with significant differences (Figure 6C). The potential metabolic function of the intestinal microbiota is caused by changes in bacterial classification. The classification information was linearly predicted based on the functional gene spectrum of microorganisms in the KEGG database. Functional prediction outcomes can be enriched at three levels of the metabolic pathways, which are represented by bars in Figure 6D–F. At level 1 of each group, the abundance of Metabolic was the highest and the abundance of Organismal systems was the lowest. At level 2 of each group, the abundance of Carbohydrate metabolism is the highest, followed by Membrane transport and Amino acid metabolism. The abundances of Metabolism of cofactors and vitamins, Nucleoside metabolism, Signal transduction, Energy metabolism and Translation are similar. Among the levels 3 of each group, the abundance of ABC transporters was the highest, followed by Two-component system and Purine metabolism. Aminoacyl-tRNA biosynthesis and Starch and sucrose metabolism in diverse environments have little difference in abundance. In contrast, Pyrimidine metabolism, Amino sugar and nucleotide sugar metabolism, and Ribosome have little difference in abundance. The results showed that compared with CG and HNG, the abundance of Metabolism at level 1, at level 2 and Metabolic pathways at level 3 increases to some extent in the CHG and CLG. It has been hypothesized that CILF can improve HN by increasing metabolic pathways associated with metabolic regulation.
Network Construction and Analysis of the Effect of CILF on HN
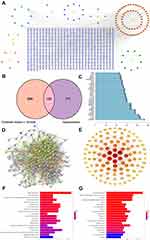
To explore the potential ingredients and molecular targets of CILF that affect HN, we conducted a network pharmacological analysis. For a comprehensive identification of the pharmacological mechanism of CILF on HN, we constructed a network between CILF active components, the corresponding targets, and disease-related genes (Figure 7A).
We identified 105 potentially active compounds and 849 gene targets corresponding to the potentially active compounds in CILF. We also found 861 related disease target genes. A Venn intersection target diagram was established, and 150 common targets were identified (Figure 7B). By node degree statistics, we found that targets such as IL-6, TNF, IL-1β, STAT3, VEGFA, and SIRT1 were located in the central network, and some markers were also considered targets in our in vivo study (Figure 7C). In combination with high-confidence node analysis in the PPI network, the core targets of the PPI network were determined by Cytoscape software (Figure 7D and E).
To better understand the functions of CILF, GO function and KEGG pathway analysis of the above 150 co-targets were used to identify the relevant pathways and functions based on the putative targets. And a series of 131 remarkably enriched GO terms were identified (p < 0.05). The top 20 terms were displayed in Figure 7F and Table S3. Furthermore, 161 KEGG pathways were identified to the requirement of p < 0.05, and bar plots of the top 20 KEEG signaling pathways were built based on p values, in which IL17 signaling pathway, TNF signaling pathway, advanced glycation end products, and receptor of glycation end products (AGE-RAGE) signaling pathway were identified as the leading pathways (Figure 7G and Table S4). These results are useful in better understanding the mechanisms of CILF in HN.
Molecular Docking Analysis
The docking analysis and binding interactions between the five potential quality markers and their connected candidate targets (IL-6, TNF, IL1-β, STAT3, VEGFA and SIRT1) were carried out using the AutoDock Vina platform. As summarized in Table S5, the binding energies were computed to evaluate the binding affinities of the five markers with their receptors. It is generally believed that ligand-receptor pairs with lower-energy binding conformational stability have a higher possibility of interaction.
All of the marker compounds showed relatively low binding free energies with their receptors, ranging from −6.3 to −8.9 kcal/mol. As a result, five molecule-target pairs, including 3’-hydroxyflavanone/SIRT1, exhibited the lowest energies (−8.9 kcal/mol). Diphenylhydantoic acid/SIRT1 also displayed the lowest binding energy (−8.9 kcal/mol), diphenylhydantoic acid/TNF exhibited the best affinity (−8.9 kcal/mol), 3’-hydroxyflavanone/TNF had the lowest energy (−8.6 kcal/mol), and metformin/TNF also showed the best affinity (−8.6 kcal/mol). The 3D and 2D molecular docking images of the nine molecule-target pairs are shown in Figure 8.
Immunohistochemistry Analysis
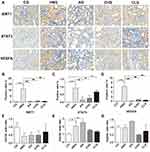
We further explored the effect of CILF on markers of renal indicators of pathological injuries. The results of the immunohistochemical staining analysis indicated that the protein expression of STAT3, VEGFA and SIRT1 was significantly increased (p < 0.001) in the HNG compared with that in the CG. Additionally, compared with HNG, the upregulation of the expression of STAT3, VEGFA and SIRT1 in renal tissue was blocked significantly by treatment with CILF or allopurinol. The results are illustrated in Figure 9A–D.
Effects of CILF on the Expression of Fibrosis Regulators in the Renal Tissues of HN Rats
To validate the protective effects of CILF on kidney damage as predicted by network pharmacology and molecular docking analysis, we measured the mRNA expression of renal fibrosis regulator factors by RT-qPCR (Sirt1, Stat3 and Vegfa). As shown in Figure 9E–G, the expression levels of Stat3 (p < 0.05 for mRNA) and Vegfa mRNA in kidney tissues were significantly increased in the HNG compared with the CG. Moreover, CILF administration significantly inhibited the expression of Stat3 and Vegfa mRNA compared with the HNG.
Discussion
It has been reported that HN, as the main complication of hyperuricemia, is due to too much UA in the body or reduced UA excreted by the kidney. Urate reaches a supersaturated state and deposits in the renal interstitium and ureter, resulting in renal lesions.38 Adenine and ethambutol contributed to the pathogenesis of urate nephropathy and HN in an animal model. In the present study, HN rats developed systematic and serious histopathological damage to the kidney, as well as tubular ectasia with UA crystal deposition and tubular interstitial inflammatory cell infiltration, along with elevated serum UA, CREA and UREA concentrations.26 Administration of low and high doses of CILF significantly reduced the increases in UA, UREA and CREA in HN rats and reduced structural damage to the kidney tissue. These results suggest that CILF can improve HN symptoms by inhibiting abnormal changes in serological characteristics and blocking the structural damage to the renal tissue.
There is no exact name for UA nephropathy in TCM. It is recorded as “Lijie disease” in the Danxi Heart Method. Hyperuricemia kidney damage is caused by “turbid blood stasis” of the kidney, which is consistent with the viewpoint of “Chronic Diseases Transforming to Collaterals” in TCM. According to its characteristics and clinical manifestations, it can be classified into the categories of “Bi symptoms”, “drenching syndrome”, “gout”, “edema”, “asthenia and fatigue”. In the early stage of UA nephropathy, deficiency of the spleen and stomach and accumulation of phlegm and dampness is more common. In the later stage, the disease is located in the spleen and kidney, mainly phlegm and blood stasis blocking collaterals, mixed with deficiency and excess.
The effects of each herb in CILF may provide theoretical insights into its effects on hyperuricemia and HN. As a component of CILF, Cichorium intybus L. is a traditional Uygur folk medicine that has the functions of clearing the liver and bile, enhancing the stomach and promoting digestion. Its pharmacological effects include antioxidant, anti-inflammatory, hyperglycemia and hyperlipidaemia. It has been reported that this herb has a good UA-lowering effect.39 The study revealed the renal protective effect of Chicory in alleviating hyperuricemia induced by high purine diet by regulating intestinal microbiota.40 Previous studies have shown that Gardenia jasminoides Ellis can regulate hyperuricemia, and gardenia fruit extract can improve hyperuricemia with renal insufficiency.41 Pueraria lobata (Willd.) Ohwi showed anti-inflammatory activity, decreased ankle swelling and alleviated pathological injury in gouty arthritis model animals.42 Similarly, Lilium brownii F.E. Brown var. viridulum Baker, a traditional herb from China, has been reported to antioxidant and anti-inflammatory, hypoglycemic and antibacterial activities.18 In addition, Morus alba L. and its extract improve diabetes related renal function injury in rats. Imperatorin, the active ingredient of Angelica Dahurica (Fisch, Ex Hoffm.) Benth. et hook.f., showed antihypertensive and antioxidant effects in renal injury of renovascular hypertensive rats.43 Taraxacum officinale Weber and its sterols have therapeutic effects on diabetes induced renal injury through in vitro and in vivo studies.44 CILF is a representative prescription consisting of seven herbs and it is clinically used to treat patients with gout and hyperuricemia in TCM. Although the individual drugs in CILF have various effects, such as anti-inflammatory, antioxidant45 and hyperuricemia effects,41 as demonstrated by in experiments, we revealed that CILF significantly decreased UA and damage in kidney tissues. In UHPLC-MS analysis, the main compounds of CILF are identified, helping to control the quality of CILF and facilitating further research, such as pharmacodynamic studies.
Although some adverse reactions may be caused by its long-term use, allopurinol is the most common classical drug used in the clinical treatment of HN.8 In our study, CILF and allopurinol showed a similar therapeutic effect on the adenine- and ethambutol-induced HN rats, which provides valid evidence that CILF is a good candidate for treating HN as an alternative to allopurinol in clinics.
Based on the gut-renal axis theory, TCM is designed to alleviate kidney damage by reducing the production of metabolites such as UA, CREA and UREA, inhibiting the accumulation of toxins leading to intestinal flora disorders and impairment of intestinal barrier function into the blood circulation, preventing the activation of severe metabolic endotoxemia and systemic inflammation, and then delay kidney damage by accelerating the excretion of toxins in the body.46 The gut microbiota is a complex ecosystem that is considered the second genome of the human body and plays an important role in supporting human health. The gut microbiota is established during early childhood development, and its composition and metabolism are closely related to chronic metabolic syndromes such as gout, diabetes, hyperuricemia and so on.47 Two-thirds of UA formed in the body is excreted through the kidneys, and one-third is excreted through the gut.48 Due to in-depth research on sequencing technology and metabolism in recent years, the correlation between gut microbiota and HN has become more obvious.22 It is not hard to imagine that the intestinal contribution of UA excretion is an important alternative pathway during renal dysfunction. Increased intestinal excretion of UA may account for the absence of significant hyperuricemia during chronic renal failure. Currently, it is believed that UA can induce kidney damage by activating inflammatory pathways, which may be one of the underlying mechanisms. The treatment of gut microbiota in HN is a hotspot of current research and a potential new treatment method in the future, which is of great significance to human health. Alterations of the gut microbiota have been previously proposed to be tightly linked to inflammatory factors in CKD.49 Moreover, the pathogenesis of HN is related to inflammation and the gut microbiota, and CILF can alter the microbial composition. After verifying its ability to alleviate HN, the effect of CILF on microbiota was investigated.
Cichorium intybus L., the main drug in CILF, has been recorded as having anti-inflammatory and antioxidant effects and preventing chronic diseases by maintaining the gut microbiota structure.15 The results of multivariate analysis showed that CILF could regulate flora structures, especially at the phylum, family and genus levels. Some studies have pointed out that the main product of Gram-negative gut bacteria, namely, Proteobacteria, is characteristic of gout and that the pathogenesis is due to the adverse effect of lipopolysaccharide (LPS) on renal function.50 The increased transfer of enteric-derived LPS to the kidneys through the blood circulation impairs the excretion of UA in the kidney and leads to the occurrence of HN.51 We found that at the level of the HNG microbial family, the number of Ruminococcaceae producing butyric acid in Firmicutes decreased, while the relative abundance of CHG and CLG increased significantly. Butyric acid, produced by intestinal epithelial cells, can promote the synthesis of intestinal mucus and enhance the function of the intestinal barrier.52 When the amount of butyric acid decreases, the resulting disorder of intestinal flora can produce large amounts of enteric-derived urinary toxins, resulting in intestinal mucosa and inflammation.53 The increased transfer of enteric-derived LPS to the kidneys through the blood circulation impairs the excretion of UA by the kidney and leads to the occurrence of HN.51 There may be a potential LPS interactional relationship between the intestine and the kidney. Therefore, it can be concluded that CILF treatment can prevent intestinal bacterial metastasis and indirectly protect renal function in HN rats by altering the intestinal microbiota and strengthening the intestinal barrier.
Communication between gut microbes and their hosts is based on bacterial metabolites. In addition to LPS, short-chain fatty acids (SCFAs) are also associated with the development of HN. There is evidence that SCFAs are an important energy source for maintaining the intestinal epithelium.54 SCFAs improve renal function by regulating inflammation, acting as antioxidants and participating in metabolic regulation. SCFA supplementation can significantly improve intestinal barrier function and renal function.55 In the correlation analysis, Lachnospiraceae and Ruminococcaceae showed a strong correlation with urinary toxins. These two bacteria are SCFA-producing bacteria.56 This result is consistent with the literature, suggesting that SCFAs may be beneficial to renal function. In recent years, many studies have found that the intestinal flora of HN patients is different from that of healthy people. This is mainly reflected in the decrease in the number of Lactobacillus.57 The above analysis showed that CILF intervention in HN rats can increase beneficial bacteria with SCFA-forming enzymes, such as Lactobacillus. As the most common probiotics, an increased abundance of Lactobacillus can increase the production of SCFA, improve intestinal barrier function, minimize the retention of UA in the body, reduce chronic intestinal and systemic inflammation, and protect renal function.58 Erysipelotrichaceae showed a possible correlation with the biological index, while Ruminococcaceae and Bacteroides had a negative correlation, which means that the first strain may reduce renal function, while the other two strains may improve renal function. In addition, in a recent study, Erysipelotrichaceae showed a positive correlation with urea in both the plasma and renal indices, which means that this strain may be harmful to renal function.59 Bifidobacterium, as a probiotic, can form a biofilm on the intestinal mucosa, which inhibits the proliferation of pathogenic bacteria and reduces the pH value of the intestine by producing butyric acid. In addition, Bifidobacterium can promote intestinal excretion, reduce the transfer of bacterial flora and its metabolites into the blood circulation, and have an important impact on improving chronic inflammation and endotoxemia.40 Combined with the current research results, it is inferred that CILF can reduce the levels of various inflammatory substances and promote the healthy function of the intestinal system by affecting the gut microbiota.
To further identify the pharmacodynamic mechanism of CILF in HN, we analyzed the binding activity between potential pharmacodynamic components and key targets through network pharmacology and molecular docking. Five potential compounds (Quercetin, 3’-Hydroxyflavanone, Beta-sitosterol, Methetoin, Diphenylhydantoic Acid), 6 core targets and three key pathways (IL17 signaling pathway, TNF signaling pathway, AGE-RAGE signaling pathway) were identified based on the network pharmacology and molecular docking results. We found that the active components in the network pharmacological analysis and molecular docking matched well with the functional targets. This shows that we can predict that CILF can play a molecular pharmacological role in preventing and treating HN by improving renal function by inhibiting the expression and signaling pathways of genes. Dysregulation of metabolism in hyperuricemia may eventually lead to tissue damage. Renal function injury is one of the most common clinical complications of hyperuricemia but the pathogenesis of HN has not been fully determined. TNF-α is a proinflammatory cytokine. Increased UA can induce oxidative stress and ROS production in vascular endothelial cells, and a large number of ROS can upregulate the expression of IL-6 and TNF-α.60 Previous studies have shown that UA, TNF-α, IL-6 and lipopolysaccharide levels were increased in hyperuricemia rat models by TNF signaling pathway analysis, indicating that patients with hyperuricemia are in a state of mild systemic inflammation. IL-17 is an important proinflammatory factor that plays an important role in inflammation and the immune response. IL-17 binds to its receptors and activates downstream pathways, including NF-κB and MAPK, thereby inducing the proinflammatory cytokines IL-6 and TNF-α and, subsequently, inflammation. These results suggest that CILF can block inflammatory and UA activities and reduce the expression of TNF-α and IL-6.61 When UA stimulated human umbilical vein endothelial cells, HMGB1 promoted its release from endothelial cells through a positive feedback mechanism. After binding to the receptor of advanced glycation end products (RAGE), UA-induced HMGB1 activates the NF-κB signaling pathway and promotes the production of the inflammatory cytokines IL-6 and TNF-α, leading to oxidative stress and inflammatory responses.62 In addition, a recent report showed that the microbiota can induce the production of IL-1β and that stimulation of IL-1β-IL-1R signaling is essential to promote the differentiation of Th17 cell.63 Studies have shown that the differentiation of cytokines related to IL-17 signaling pathway is related to the colonization of intestinal flora and may participate in intestinal immune homeostasis.64
Interestingly, we also found consistent results between the in vivo studies and the network pharmacology analysis and molecular docking predictions. RT-qPCR and immunohistochemistry analysis further validated some possible mechanisms by which CILF, inhibits renal UA metabolism and protects renal function, as well as ameliorating changes in inflammation in HN rat kidney tissue. VEGFA is significantly increased in patients with acute kidney injury. In addition, miR-195-5p prevents acute kidney injury by targeting VEGFA to inhibit inflammation and oxidative stress.65 A large amount of evidence has shown that STAT3 is one of the main signaling pathways involved in the progression of renal fibrosis. Pharmacological blockade of STAT3 delayed the progression of renal fibrosis in hyperuricemia-induced CKD. Treatment with the STAT3 inhibitor S3I-201 improved renal insufficiency, reduced serum UA levels, and delayed the progression of renal fibrosis.66 SIRT1 is a histone deacetylase associated with a variety of biological processes in several organisms. In the kidney, SIRT1 inhibits renal cell apoptosis, inflammation and fibrosis and can reduce UA levels and protect the kidney by inhibiting the activity of nuclear transcription factors and inflammasomes.67 In this study, we found increased expression of VEGFA, STAT3, and SIRT1 in the renal tissues of HN rats, and these increases were significantly inhibited by CILF administration.
We identified the stable existence of chemical substances both in CILF and each single component herb to ensure the quality of the CILF. Our study on the classical pathways of CILF in HN identified by network pharmacology and molecular docking technology plus an analysis of gut microbiota can offer further information for understanding the function of potent chemicals and mechanisms in vivo, such as metabonomic analysis. In the future, our group will conduct in-depth research to solve these problems to provide a new approach to the prevention and treatment of HN.
Conclusion
Treatment of adenine and ethambutol-induced HN rats with CILF for 3 weeks improved their serological imbalance and renal injury. By applying an analysis of their gut microbiota, network pharmacology, and molecular docking techniques, CILF has been proven to alter the gut bacterial diversity and community structure and to improve HN by regulating the gut microbiota. CILF can play a molecular pharmacological role in improving renal function by inhibiting the expression and signaling pathways of genes. Our results show that CILF has a beneficial role in the clinical treatment and prevention of HN progression in patients with hyperuricemia.
Abbreviations
AGE-RAGE, advanced glycation end products, and receptor of glycation end products; CILF, Cichorium intybus L. formula; CKD, chronic kidney disease; CREA, creatinine; CV, coefficient of variation; DL, drug-likeness; GO, gene ontology; H&E, hematoxylin and eosin; HN, hyperuricemic nephropathy; KEGG, Kyoto encyclopedia of genes and genomes; LefSe, LDA effect size; LPS, lipopolysaccharide; NMDS, non-metric multidimensional scaling; OB, oral bioavailability; OTUs, operational taxonomic units; PCoA, principal coordinates analysis; PD, faith’s phylogenetic diversity; RT-qPCR, real-time quantitative polymerase chain reaction; SIRT1, Sirtuin 1; STAT3, signal transducers and activators of transduction-3; TCM, traditional Chinese medical; UA, uric acid; UREA, serum urea nitrogen; VEGFA, vascular endothelial growth factor-A.
Data Sharing Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/; 16S rRNA sequencing data is available via BioProject PRJNA838749.
Ethics Statement
The study was reviewed and approved by the Ethics Committee of School of Pharmacy of Southwest Minzu University (Approval No. 2022-09).
Acknowledgments
The author thanks Chengdu Lilai Biomedicine Experiment Center for the technical support for immunohistochemistry.
Funding
This work was supported by National Natural Science Foundation of China (No. 81801086); Natural Science Foundation of Sichuan, China (No.2022NSFSC1574) and the innovative research project for graduate students of Southwest Minzu University in 2022 (No. ZD2022168).
Disclosure
The authors declare that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
References
1. Isaka Y, Takabatake Y, Takahashi A, Saitoh T, Yoshimori T. Hyperuricemia-induced inflammasome and kidney diseases. Nephrol Dial Transplant. 2016;31(6):890–896. doi:10.1093/ndt/gfv024
2. Yokozawa T, Zheng PD, Oura H, Koizumi F. Animal model of adenine-induced chronic renal failure in rats. Nephron. 1986;44(3):230–234. doi:10.1159/000183992
3. Postlethwaite AE, Kelley WN. Studies on the mechanism of ethambutol-induced hyperuricemia. Arthritis Rheum. 1972;15(4):403–409. doi:10.1002/art.1780150411
4. Meng Z, Yan Y, Tang Z, et al. Anti-hyperuricemic and nephroprotective effects of rhein in hyperuricemic mice. Planta Med. 2015;81(4):279–285. doi:10.1055/s-0034-1396241
5. Feng Y, Sun F, Gao Y, et al. Taurine decreased uric acid levels in hyperuricemic rats and alleviated kidney injury. Biochem Biophys Res Commun. 2017;489(3):312–318. doi:10.1016/j.bbrc.2017.05.139
6. Li Y, Zhao Z, Luo J, et al. Apigenin ameliorates hyperuricemic nephropathy by inhibiting URAT1 and GLUT9 and relieving renal fibrosis via the Wnt/beta-catenin pathway. Phytomedicine. 2021;87:153585. doi:10.1016/j.phymed.2021.153585
7. Kwak CH, Sohn M, Han N, Cho YS, Kim YS, Oh JM. Effectiveness of febuxostat in patients with allopurinol-refractory hyperuricemic chronic kidney disease. Int J Clin Pharmacol Ther. 2018;56(7):321–327. doi:10.5414/CP202735
8. Kabul S, Shepler B. A review investigating the effect of allopurinol on the progression of kidney disease in hyperuricemic patients with chronic kidney disease. Clin Ther. 2012;34(12):2293–2296. doi:10.1016/j.clinthera.2012.10.008
9. Fujimori S, Ooyama K, Ooyama H, Moromizato H. Efficacy of benzbromarone in hyperuricemic patients associated with chronic kidney disease. Nucleosides Nucleotides Nucleic Acids. 2011;30(12):1035–1038. doi:10.1080/15257770.2011.622732
10. Wang H, Peng Y, Zhang T, et al. Metabolic epoxidation is a critical step for the development of benzbromarone-induced hepatotoxicity. Drug Metab Dispos. 2017;45(12):1354–1363. doi:10.1124/dmd.117.077818
11. Bardin T. Current management of gout in patients unresponsive or allergic to allopurinol. Joint Bone Spine. 2004;71(6):481–485. doi:10.1016/j.jbspin.2004.07.006
12. Xiao E, Luo L. Alternative therapies for diabetes: a comparison of Western and traditional Chinese medicine (TCM) approaches. Curr Diabetes Rev. 2018;14(6):487–496. doi:10.2174/1573399813666170519103230
13. Huijuan W, Xiaoxu C, Rui S, Xinghui L, Beibei T, Jianchun M. Qi-Zhu-Xie-Zhuo-Fang reduces serum uric acid levels and ameliorates renal fibrosis in hyperuricemic nephropathy rats. Biomed Pharmacother. 2017;91:358–365. doi:10.1016/j.biopha.2017.04.031
14. Pan J, Shi M, Ma L, Fu P. Mechanistic insights of soluble Uric acid-related kidney disease. Curr Med Chem. 2020;27(30):5056–5066. doi:10.2174/0929867326666181211094421
15. Janda K, Gutowska I, Geszke-Moritz M, Jakubczyk K. The common cichory (Cichorium intybus L.) as a source of extracts with health-promoting properties-A review. Molecules. 2021;26(6):1814. doi:10.3390/molecules26061814
16. Chen L, Li M, Yang Z, et al. Gardenia jasminoides Ellis: ethnopharmacology, phytochemistry, and pharmacological and industrial applications of an important traditional Chinese medicine. J Ethnopharmacol. 2020;257:112829. doi:10.1016/j.jep.2020.112829
17. Zhou YX, Zhang H, Peng C. Puerarin: a review of pharmacological effects. Phytother Res. 2014;28(7):961–975. doi:10.1002/ptr.5083
18. Zhou J, An R, Huang X. Genus Lilium: a review on traditional uses, phytochemistry and pharmacology. J Ethnopharmacol. 2021;270:113852. doi:10.1016/j.jep.2021.113852
19. Liang WH, Chang TW, Charng YC. Influence of harvest stage on the pharmacological effect of Angelica dahurica. Bot Stud. 2018;59(1):14. doi:10.1186/s40529-018-0230-1
20. Ma G, Chai X, Hou G, Zhao F, Meng Q. Phytochemistry, bioactivities and future prospects of mulberry leaves: a review. Food Chem. 2022;372:131335. doi:10.1016/j.foodchem.2021.131335
21. Gonzalez-Castejon M, Visioli F, Rodriguez-Casado A. Diverse biological activities of dandelion. Nutr Rev. 2012;70(9):534–547. doi:10.1111/j.1753-4887.2012.00509.x
22. Yang T, Richards EM, Pepine CJ, Raizada MK. The gut microbiota and the brain-gut-kidney axis in hypertension and chronic kidney disease. Nat Rev Nephrol. 2018;14(7):442–456. doi:10.1038/s41581-018-0018-2
23. Evenepoel P, Poesen R, Meijers B. The gut-kidney axis. Pediatr Nephrol. 2017;32(11):2005–2014. doi:10.1007/s00467-016-3527-x
24. Zhang RZ, Yu SJ, Bai H, Ning K. TCM-Mesh: the database and analytical system for network pharmacology analysis for TCM preparations. Sci Rep. 2017;7(1):2821. doi:10.1038/s41598-017-03039-7
25. Li N, Amatjan M, He P, et al. Integration of network pharmacology and intestinal flora to investigate the mechanism of action of Chinese herbal Cichorium intybus formula in attenuating adenine and ethambutol hydrochloride-induced hyperuricemic nephropathy in rats. Pharm Biol. 2022;60(1):2338–2354. doi:10.1080/13880209.2022.2147551
26. Wu H, Zhou M, Lu G, Yang Z, Ji H, Hu Q. Emodinol ameliorates urate nephropathy by regulating renal organic ion transporters and inhibiting immune inflammatory responses in rats. Biomed Pharmacother. 2017;96:727–735. doi:10.1016/j.biopha.2017.10.051
27. Duan S, Niu L, Yin T, et al. A novel strategy for screening bioavailable quality markers of traditional Chinese medicine by integrating intestinal absorption and network pharmacology: Application to Wu Ji Bai Feng Pill. Phytomedicine. 2020;76:153226. doi:10.1016/j.phymed.2020.153226
28. Guo X, Wu Y, Zhang C, Wu L, Qin L, Liu T. Network Pharmacology Analysis of ZiShenWan for Diabetic Nephropathy and Experimental Verification of Its Anti-Inflammatory Mechanism. Drug Des Devel Ther. 2021;15:1577–1594. doi:10.2147/DDDT.S297683
29. Ye Q, Zhang Q, Yao H, et al. Active-ingredient screening and synergistic action mechanism of shegan mixture for anti-asthma effects based on network pharmacology in a mouse model of Asthma. Drug Des Devel Ther. 2021;15:1765–1777. doi:10.2147/DDDT.S288829
30. Meng X, Ma J, Kang AN, Kang SY, Jung HW, Park YK. A novel approach based on metabolomics coupled with intestinal flora analysis and network pharmacology to explain the mechanisms of action of bekhogainsam decoction in the improvement of symptoms of streptozotocin-induced diabetic nephropathy in mice. Front Pharmacol. 2020;11:633. doi:10.3389/fphar.2020.00633
31. Tong H, Yu M, Fei C, et al. Bioactive constituents and the molecular mechanism of Curcumae Rhizoma in the treatment of primary dysmenorrhea based on network pharmacology and molecular docking. Phytomedicine. 2021;86:153558. doi:10.1016/j.phymed.2021.153558
32. Yin K, Li X, Luo X, et al. Hepatoprotective effect and potential mechanism of aqueous extract from phyllanthus emblica on carbon-tetrachloride-induced liver fibrosis in rats. Evid Based Complement Alternat Med. 2021;2021:5345821. doi:10.1155/2021/5345821
33. Jhang JJ, Lin JH, Yen GC. Beneficial properties of phytochemicals on NLRP3 inflammasome-mediated gout and complication. J Agric Food Chem. 2018;66(4):765–772. doi:10.1021/acs.jafc.7b05113
34. Tan J, Wan L, Chen X, et al. Conjugated linoleic acid ameliorates high fructose-induced hyperuricemia and renal inflammation in rats via NLRP3 Inflammasome and TLR4 signaling pathway. Mol Nutr Food Res. 2019;63(12):e1801402. doi:10.1002/mnfr.201801402
35. Loomba R, Seguritan V, Li W, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017;25(5):1054–1062 e5. doi:10.1016/j.cmet.2017.04.001
36. Zhao L, Zhang F, Ding X, et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science. 2018;359(6380):1151–1156. doi:10.1126/science.aao5774
37. Dubin K, Callahan MK, Ren B, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. doi:10.1038/ncomms10391
38. Park JH, Jo YI, Lee JH. Renal effects of uric acid: hyperuricemia and hypouricemia. Korean J Intern Med. 2020;35(6):1291–1304. doi:10.3904/kjim.2020.410
39. Bian M, Lin Z, Wang Y, Zhang B, Li G, Wang H. Bioinformatic and Metabolomic Analysis Reveal Intervention Effects of Chicory in a Quail Model of Hyperuricemia. Evid Based Complement Alternat Med. 2018;2018:5730385. doi:10.1155/2018/5730385
40. Bian M, Wang J, Wang Y, et al. Chicory ameliorates hyperuricemia via modulating gut microbiota and alleviating LPS/TLR4 axis in quail. Biomed Pharmacother. 2020;131:110719. doi:10.1016/j.biopha.2020.110719
41. Hu QH, Zhu JX, Ji J, Wei LL, Miao MX, Ji H. Fructus Gardenia Extract ameliorates oxonate-induced hyperuricemia with renal dysfunction in mice by regulating organic ion transporters and mOIT3. Molecules. 2013;18(8):8976–8993. doi:10.3390/molecules18088976
42. Wang X, Zhang Y, Zhang M, et al. Novel carbon dots derived from puerariae lobatae radix and their anti-gout effects. Molecules. 2019;24(22):4152. doi:10.3390/molecules24224152
43. Cao YJ, He X, Wang N, He LC. Effects of imperatorin, the active component from radix angelicae (Baizhi), on the blood pressure and oxidative stress in 2K,1C hypertensive rats. Phytomedicine. 2013;20(12):1048–1054. doi:10.1016/j.phymed.2013.04.021
44. Tian L, Fu P, Zhou M, Qi J. Dandelion sterol improves diabetes mellitus-induced renal injury in in vitro and in vivo study. Food Sci Nutr. 2021;9(9):5183–5197. doi:10.1002/fsn3.2491
45. Nwafor IC, Shale K, Achilonu MC. Chemical composition and nutritive benefits of chicory (Cichorium intybus) as an ideal complementary and/or alternative livestock feed supplement. ScientificWorldJournal. 2017;2017:7343928. doi:10.1155/2017/7343928
46. Xu X, Wang H, Guo D, et al. Curcumin modulates gut microbiota and improves renal function in rats with uric acid nephropathy. Ren Fail. 2021;43(1):1063–1075. doi:10.1080/0886022X.2021.1944875
47. Xu J, Lian F, Zhao L, et al. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 2015;9(3):552–562. doi:10.1038/ismej.2014.177
48. Buzard J, Bishop C, Talbott JH. Recovery in humans of intravenously injected isotopic uric acid. J Biol Chem. 1952;196(1):179–184. doi:10.1016/S0021-9258(18)55717-3
49. Li F, Wang M, Wang J, Li R, Zhang Y. Alterations to the gut microbiota and their correlation with inflammatory factors in chronic kidney disease. Front Cell Infect Microbiol. 2019;9:206. doi:10.3389/fcimb.2019.00206
50. Liu X, Lv Q, Ren H, et al. The altered gut microbiota of high-purine-induced hyperuricemia rats and its correlation with hyperuricemia. PeerJ. 2020;8:e8664. doi:10.7717/peerj.8664
51. Johansson ME, Hansson GC. Microbiology. Keeping bacteria at a distance. Science. 2011;334(6053):182–183. doi:10.1126/science.1213909
52. Feng W, Ao H, Peng C. Gut microbiota, short-Chain fatty acids, and herbal medicines. Front Pharmacol. 2018;9:1354. doi:10.3389/fphar.2018.01354
53. Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, Macia L. The role of short-chain fatty acids in health and disease. Adv Immunol. 2014;121:91–119. doi:10.1016/B978-0-12-800100-4.00003-9
54. Chen R, Xu Y, Wu P, et al. Transplantation of fecal microbiota rich in short chain fatty acids and butyric acid treat cerebral ischemic stroke by regulating gut microbiota. Pharmacol Res. 2019;148:104403. doi:10.1016/j.phrs.2019.104403
55. Wei Z, Cui Y, Tian L, et al. Probiotic Lactiplantibacillus plantarum N-1 could prevent ethylene glycol-induced kidney stones by regulating gut microbiota and enhancing intestinal barrier function. FASEB J. 2021;35(11):e21937. doi:10.1096/fj.202100887RR
56. Zhao L, Lou H, Peng Y, Chen S, Zhang Y, Li X. Comprehensive relationships between gut microbiome and faecal metabolome in individuals with type 2 diabetes and its complications. Endocrine. 2019;66(3):526–537. doi:10.1007/s12020-019-02103-8
57. Gao Y, Sun J, Zhang Y, et al. Effect of a traditional Chinese medicine formula (CoTOL) on serum uric acid and intestinal flora in obese hyperuricemic mice inoculated with intestinal bacteria. Evid Based Complement Alternat Med. 2020;2020:8831937. doi:10.1155/2020/8831937
58. Wanchai K, Yasom S, Tunapong W, et al. Probiotic Lactobacillus paracasei HII01 protects rats against obese-insulin resistance-induced kidney injury and impaired renal organic anion transporter 3 function. Clin Sci (Lond). 2018;132(14):1545–1563. doi:10.1042/CS20180148
59. Liu S, Liu H, Chen L, et al. Effect of probiotics on the intestinal microbiota of hemodialysis patients: a randomized trial. Eur J Nutr. 2020;59(8):3755–3766. doi:10.1007/s00394-020-02207-2
60. Zhou Y, Zhao M, Pu Z, Xu G, Li X. Relationship between oxidative stress and inflammation in hyperuricemia: analysis based on asymptomatic young patients with primary hyperuricemia. Medicine. 2018;97(49):e13108. doi:10.1097/MD.0000000000013108
61. Liu P, Xu H, Shi Y, Deng L, Chen X. Potential Molecular Mechanisms of Plantain in the Treatment of Gout and Hyperuricemia Based on Network Pharmacology. Evid Based Complement Alternat Med. 2020;2020:3023127. doi:10.1155/2020/3023127
62. Cai W, Duan XM, Liu Y, et al. Uric Acid Induces Endothelial Dysfunction by Activating the HMGB1/RAGE Signaling Pathway. Biomed Res Int. 2017;2017:4391920. doi:10.1155/2017/4391920
63. Shaw MH, Kamada N, Kim YG, Nunez G. Microbiota-induced IL-1beta, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J Exp Med. 2012;209(2):251–258. doi:10.1084/jem.20111703
64. Wang Y, Yin Y, Chen X, et al. Induction of intestinal Th17 cells by flagellins from segmented filamentous bacteria. Front Immunol. 2019;10:2750. doi:10.3389/fimmu.2019.02750
65. Xu Y, Jiang W, Zhong L, et al. miR-195-5p alleviates acute kidney injury through repression of inflammation and oxidative stress by targeting vascular endothelial growth factor A. Aging. 2020;12(11):10235–10245. doi:10.18632/aging.103160
66. Pan J, Shi M, Guo F, Ma L, Fu P. Pharmacologic inhibiting STAT3 delays the progression of kidney fibrosis in hyperuricemia-induced chronic kidney disease. Life Sci. 2021;285:119946. doi:10.1016/j.lfs.2021.119946
67. Chen L, Lan Z. Polydatin attenuates potassium oxonate-induced hyperuricemia and kidney inflammation by inhibiting NF-kappaB/NLRP3 inflammasome activation via the AMPK/SIRT1 pathway. Food Funct. 2017;8(5):1785–1792. doi:10.1039/c6fo01561a
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.