Back to Journals » OncoTargets and Therapy » Volume 13
A New Fusion Peptide Targeting Pancreatic Cancer and Inhibiting Tumor Growth
Authors Zheng L, Zhang B , He X, Cao G, Li Y, Cai K, Yang B, Wu Y
Received 22 January 2020
Accepted for publication 26 July 2020
Published 12 August 2020 Volume 2020:13 Pages 7865—7875
DOI https://doi.org/10.2147/OTT.S246969
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Lei Zheng,1 Bo Zhang,1,2 Xiaoman He,1 Guodong Cao,1 Yongzhou Li,1 Kailun Cai,1 Bin Yang,1,2 Yulian Wu1,2
1Department of Surgery, The Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310000, People’s Republic of China; 2Key Laboratory of Cancer Prevention and Intervention, China National Ministry of Education, Cancer Institute, Second Affiliated Hospital, Zhejiang University School of Medicine, Hangzhou 310000, People’s Republic of China
Correspondence: Yulian Wu Email [email protected]
Background: Pancreatic cancer is a highly malignant tumor of the digestive system. Early pancreatic cancer is often difficult to diagnosis due to its atypical clinical symptoms. Patients with pancreatic cancer have a very poor prognosis because they have lost the opportunity for radical surgical tumor resection and they are less sensitive to the clinically used radiotherapy and chemotherapy.
Methods: In this study, a peptide targeting pancreatic cancer cells was screened by phage display technology, and its targeting property was evaluated in vitro using PANC1 cells by fluorescence imaging and flow cytometry. Furthermore, the targeting peptide was conjugated to the pro-apoptotic KLAKLAKKLAKLAK (KLA), the fusion peptide and its targeting ability that allowing KLA to specifically enter pancreatic tumor cells in vitro and in vivo was confirmed by fluorescence imaging and in vivo imaging system (IVIS). Its mechanism was determined using flow cytometry, mitochondrial membrane potential evaluation and Western blot. The inhibitory effect on pancreatic tumor growth and toxic effects were evaluated by animal experiment.
Results: Due to the internalization facilitated by the targeting mechanism of the targeting peptide, KLA specifically entered pancreatic cancer cells, destroyed mitochondria and induced apoptosis. The fusion peptide and its targeting ability that allowing KLA to specifically enter pancreatic tumor cells and exert a significant inhibitory effect on pancreatic tumor growth with reduced toxic effects.
Conclusion: This approach possesses potential advantages in the clinical diagnosis and treatment of pancreatic cancer.
Keywords: targeting peptide, phage display technology, pancreatic cancer, KLA pro-apoptotic peptide, tumor growth inhibition
Introduction
Pancreatic cancer is one of the most common malignant tumors of the digestive system. It is characterized by late clinical findings, high degree of malignancy and extremely poor prognosis.1 Radical surgery is the first choice in the treatment of pancreatic cancer, but early pancreatic cancer is often difficult to diagnosis due to atypical clinical symptoms. Most of the clearly diagnosed pancreatic cancer is in its advanced stage, and 80% of patients have lost the chance of radical surgical resection.2 Moreover, pancreatic cancer has a high degree of malignancy and is poorly sensitive to clinically used radiotherapy and chemotherapy. Therefore, the overall survival rate of patients in 5 years is less than 10%.3,4 Thus, improving the diagnosis of early pancreatic cancer and the sensitivity to chemotherapeutic drugs has been the focus of our present research.
Currently, early screening for pancreatic cancer relies mainly on imaging and molecular markers, such as ultrasound, computed tomography, magnetic resonance cholangiopancreatography and endoscopic ultrasound.5 Compared with imaging examinations and more invasive histological examinations, tumor markers have the advantage of an easy serum collection, small trauma, and economical convenience.6 The most commonly used tumor marker for an early screening of pancreatic cancer is the carbohydrate antigen 19–9 (CA19-9). However, studies showed that CA19-9 still has some limitations in the early screening of pancreatic cancer due to lack of specificity,7 which currently represents still a problem because of the lack of specific early screening markers for this cancer type.
In terms of treatment, surgery is currently the only approach that can significantly prolong the survival time of patients, although overall, the rate of surgical resection in various countries is not high.8 Gemcitabine-based chemotherapy for the advanced treatment of pancreatic cancer can prolong the overall survival of patients in targeted therapy.9 However, chemotherapeutic drugs still have tremendous toxic effects on the patient’s body.
Therefore, the goal of this work was to develop a new peptide that specifically target pancreatic cancer cells for clinical diagnostic imaging and for treatment. Peptides that target pancreatic cancer cells PANC-1 were firstly screened using phage display technology, which is a powerful approach for the generation of peptides and antibodies that target specific organ or tumor structures.10 This technology uses genetic engineering to insert foreign gene fragments into a specific location of the gene, maintaining the relative spatial structure and biological activity of the recombinant fusion protein by expressing the protein or polypeptide encoded by the foreign gene on the surface of the phage.11 Our work demonstrated that HMNPWSD peptide was able to target panc1 cancer cells and tumors in vitro and in vivo.
KLA peptide was used as a therapeutic peptide. Compared with chemotherapeutic drugs, they offer unique advantages, such as small molecular weight, tumor-penetrating ability, low immunogenicity, good biocompatibility, lower toxic effects, and ease of synthesis and modification.12,13 KLA, a typical amphiphilic a-helical pro-apoptotic peptide, with the following amino acid sequence of D(KLAKLAK)2, induces apoptosis in mammalian cells via the disruption of the mitochondrial membrane.14,15 However, the KLA peptide itself cannot cross the cell membrane and cannot disrupt the mitochondrial membrane to trigger apoptosis.16 However, after the conjugation of KLA with the HMNPWSD peptide, KLA could target PANC-1 cells, enter the PANC-1 cells across the cell membrane, destroy mitochondria and induce apoptosis with the internalization facilitated by HMNPWSD. The fusion peptide has a significant inhibitory effect on PANC-1 cell proliferation and tumor growth with less toxicity on cells and animals. These findings demonstrated that HMNPWSD, in combination with KLA, had a great potential in the diagnosis and treatment of pancreatic cancer.
Materials and Methods
Cell Lines and Cell Culture
Human pancreatic cancer cell line PANC-1 and human non-small cell lung carcinoma cell line H1299 were obtained from the Academy of Sciences Cell Bank of Shanghai. PANC-1 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Shanghai, China) and H1299 cells were maintained in RPMI-1640 medium (Gibco, Shanghai, China). Both DMEM and RPMI-1640 were supplemented with 10% (v/v) FBS and 1% (v/v) penicillin-streptomycin. All cells were incubated at 37°C in a 5% CO2 atmosphere.
Biopanning
A total amount of 2×106 PANC-1 cells were cultured with 2×1011 pfu mL−1 in 10 μL M13 bacteriophage (Ph. D-C7C, NEB) in a tube that was placed on a 3D shaker for 1 h at room temperature. The supernatant was discarded and the cells were washed with 1 mL serum-free DMEM (Gibco, Shanghai, China). After four intensive washes using 1 mL PBST solution, the cell-bound phages were eluted by incubation with 600 μL BSA 0.1 wt-% in 0.2M HCl (pH 2.2) for 8 min at room temperature. Hundred and fifty μL Tris-HCl 1 M (pH 9.0) were added to neutralize the solution and the eluted phage was recovered. Phages were titrated and amplified, and the same procedure was repeated other three times. Thirty random plaques were selected from the last round. Each plaque was amplified, DNA was extracted and sent to Boshang Biotechnology (Shanghai, China) for sequencing and then the sequence of our peptide of interest was obtained.
Enzyme-Linked Immunosorbent Assay (ELISA)
PANC-1 cells and H1299 cells were separately seeded in 96-well plates at a concentration of 1x104 per well in 200 μL medium/well and placed in a CO2 incubator at 37°C for 24 h. Six selected phage clones after DNA sequencing with 2×1011 pfu mL−1 in 10 μL PBS were added to each well, and incubated for 1 h at room temperature under shaking. The unselected wild phage clone was used as a negative control. Cells were washed with PBST for 3 times, 100 μL HRP-labeled anti-M13 mAb (Sinobiological, Beijing, China) were added and cells were incubated at room temperature for 1 h. The mixture was washed with PBST 5 times, and then tetrabenzidine (SolarbioTechnology, Beijing, China) was added. Sulfuric acid 2 mol/L was used to stop the reaction. The absorbance was read at 450 nm in a microplate reader. According to the results, clone 1 and clone 2 were synthesized and conjugated with FITC by Huabio (Hangzhou, China).
In vitro Binding Assay of the Selected Peptides to Cells
To confirm the targeting specificity of the selected peptide HMNPWSD and TAPHRSL to PANC-1 cells, 1×105 PANC-1 cells were seeded into a 6-well plate; then, 1 μM HMNPWSD-FITC, TAPHRSL-FITC and SPPTGIN-FITC (control peptide) was separately added into each well, and all cells were incubated for 2 h at room temperature. H1299 cell lines were treated with 1 μM HMNPWSD-FITC under the same conditions as the control. Cells were washed with PBS three times and DAPI was added. The fluorescent signal was visualized using a fluorescence microscope.
Furthermore, flow cytometry analysis was performed to quantitatively analyze the binding affinity of the peptide to the different cell types. PANC-1 cell line and H1299 cell line were incubated with HMNPWSD-FITC at different concentrations (0, 0.5 and 1 μM) for 2 h at 37°C; then, the cells were collected and resuspended in 200 μL PBS after centrifugation. Fluorescence intensity was measured by flow cytometry.
Evaluation of HMNPWSD-KLA Binding Affinity to PANC-1 Cells in vitro and in vivo
FITC-HMNPWSD-KLA(D(KLAKLAK)2) and FITC-control-KLA were synthesized by GL Biochem (Shanghai, China). PANC-1 cells were separately incubated with 1 μM FITC-HMNPWSD-KLA and FITC-SPPTGIN-KLA for 2 h. H1299 cells were treated with 1 μM FITC-HMNPWSD-KLA at the same conditions. Cells were washed with PBS three times and DAPI was added. The fluorescent signal was visualized using a fluorescence microscope.
A total amount of 4×106 PANC-1 cells and H1299 cells in 100 μM PBS were separately subcutaneously inoculated in male nude mice of three weeks. After sixteen days the tumor grew to approximately 0.8 cm3. Then, 5 mg/kg FITC-HMNPWSD-KLA was intravenously injected into the mice carrying the PANC-1 and H1299 tumor through the tail vein. The control peptide FITC-SPPTGIN-KLA was also intravenously injected into another mouse carrying the PANC-1 tumor. The mice were sacrificed after three hours. The tumor, spleen, kidney, pancreas and liver were harvested and their fluorescence intensity was quantified by an in vivo imaging system. All animal experiments were carried out in accordance with the ethical guidelines of Animal Experimentation Committee in College of Medicine, Zhejiang University, which approved this study.
Cell Viability Assay
Cell viability was measured by the MTT assay. PANC-1 cells and H1299 cells were seeded in 96-well plates at a density of 1×104 cells per well and cultured in DMEM containing 10% FBS for 24 h. Different concentrations (0, 10, 20, 40 and 60 μM) of HMNPWSD-KLA, control-KLA, or KLA peptide were added to the wells and the cells were incubated for 24 h. Then, MTT was added and cells were incubated for 4 hours. DMSO was added after centrifugation and the color development was read at an OD of 570 nm in a microplate reader.
Mitochondrial Membrane Potential Evaluation
The decrease of mitochondrial membrane potential is a landmark event in the early stage of apoptosis. The decrease in cell membrane potential can be easily detected by the transition from red fluorescence to green fluorescence of JC-1. At the same time, the transition of red fluorescence to green fluorescence of JC-1 can also be used as a detection indicator in the early stage of apoptosis. A total of 1×105 PANC-1 and H1299 cells were separately cultured in each well of 6-well plates and incubated at 37°C for 24 h, then treated with 1 μM HMNPWSD-KLA and control peptide SPPTGIN-KLA for 2 hours, respectively. JC-1 staining working solution was prepared according to the JC-I kit instructions (Solarbio technology, Beijing, China). One mL JC-1 working solution was added to each well and cells were incubated at 37°C for 20 minutes. Then, cells were washed twice with JC-1 buffer, 2 mL PBS was added, and cells were observed under a fluorescence microscope.
Apoptosis Assay and Apoptotic Signaling Proteins
PANC-1 cells were cultured in a 6-well plate (1×105 cells/well) and incubated at 37° C for 24 h. Then they were treated with different concentrations (0, 5, 10 and 20 μM) of HMNPWSD-KLA and incubated at 37° C for 24 hours. Subsequently, cells were digested with EDTA-free enzyme, washed twice with PBS, and 500 μL binding buffer, 5 μL Annexin V-FITC, and 5 μL Propidium Iodide were added (Keygen Biotech China). The reaction was carried out for 10 minutes at room temperature in the dark. The apoptotic cells were assessed using a flow cytometer.
To further validate KLA-induced apoptosis, PANC-1 cells were treated with different concentrations (0, 5 and 10 μM) of HMNPWSD-KLA, control-KLA, or KLA and incubated at 37° C for 24 h. The PANC-1 cells were harvested, lysed with RIPA buffer, and quantified after protein extraction. The total cell proteins were separated by electrophoresis in a 10% SDS-PAGE followed by Western blot. Cells were then treated with mouse anti-human primary antibodies anti-cytochrome c, anti-cleaved caspase-3, and anti-cleaved caspase-9 (Huabio, Hangzhou, China) and incubated overnight at 4°C. Then, the secondary antibody horseradish peroxidase (HRP) conjugated goat anti mouse IgG (Huabio, Hangzhou, China) was added and cells were incubated at room temperature for 2 h prior to exposure.
Effect of Fusion Peptide on Tumor Growth and Systemic Toxicity in vivo
An amount of 4×106 PANC-1 cells were subcutaneously inoculated in male nude mice of three weeks. After sixteen days the tumor grew to approximately 0.2 cm3. Tumor-bearing mice were divided into A, B, C, D groups, each group containing 5 mice and subjected to different treatments. Five mg/kg HMNPWSD-KLA, control-KLA and KLA peptide were separately injected into the tail vein of A, B and C group, respectively, every three days for six weeks. Group D was treated with PBS as a negative control group. The tumor size was measured by a Vernier caliper and the relative tumor volume (RTV) was calculated during the treatment. The index associated to the anti-tumor activity is relative tumor growth rate (T/C (%)) according to the following formula: T/C (%) = (TRTV/CRTV) x100, in which TRTV is the relative tumor volume in the treatment group, while CRTV is the relative tumor volume in the negative control group. The efficacy evaluation criteria were as follows: T/C (%) >60 meant that the treatment was not effective; T/C (%) <60 meant that the treatment was effective. Body weights were measured every seven days during the treatment period.
In order to evaluate the toxicity of the peptide on the body, hematological parameters (WBC, RBC, PLT and HGB) and liver enzymes (ALT and AST) were measured on nude mice. The main organs were collected, stored in formalin, embedded in paraffin and 4 μm-thick slices were cut using a microtome. HE staining was performed using a standard protocol.
Results
Biopanning of Cell-Targeting Peptide for PANC-1 Cells
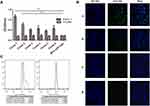
Four rounds of phage screening were performed on PANC-1 cells. The number of recovered phages increased each round (Table 1), and the third round was 67 times more enriched than the first round, indicating that the peptides targeting PANC-1 cells were effectively enriched. The fourth round of screening results showed no significant enrichment compared to the third round, indicating that the fourth round of screening tends to be saturated. After the fourth round of screening, 30 blue plaques were randomly selected and subjected to DNA extraction for sequencing. Amino acid sequence was deduced according to the brief genetic code table provided by the reagent. Based on the frequency of the sequencing results, six amino acid sequences with the highest frequency were selected and synthesized (Table 2). To evaluate the affinity of the six peptides for PANC-1 cells, ELISA was performed and the results showed that the affinity of HMNPWSD and TAPHRSL for PANC-1 cells was much higher than that of the other four peptides (Figure 1A). Thus, these two peptides were chosen for our subsequent experiments.
 |
Table 1 Enrichment of the PANC-1 Targeted Phages from Each Biopanning Round |
 |
Table 2 Amino Acid Sequence and Frequency of Thirty Random Phage Plaques Selected from the Fourth Round |
Evaluation of the Specific Binding Affinity of the Peptide-FITC Conjugate
To further evaluate the specificity of the selected HMNPWSD and TAPHRSL peptide, the selected two and the random peptide SPPTGIN were synthesized and conjugated with FITC at the N-terminus. PANC-1 cell line was treated with both the HMNPWSD-FITC and TAPHRSL-FITC, while H1299 cell line was treated with HMNPWSD-FITC as a control group. The fluorescence intensity of the HMNPWSD-FITC peptide was strong in PANC-1, but weak in H1299, while the fluorescence intensity of the TAPHRSL and control peptide SPPTGIN was weak in PANC-1 (Figure 1B). Many peptides screened by phage display technology are non-specific binding to target cells or with weak affinity which cannot be completely eliminated in the biopanning part. The number of peptide-FITC bound to cell is relatively small and show a weak signal. To quantitatively measure the binding specificity of HMNPWSD to cells, FITC fluorescence intensity was measured by flow cytometry. The results shown in Figure 1C demonstrated that the HMNPWSD peptide had a higher binding capacity to PANC-1, while no specificity to H1299 was found.
Fusion Peptide Targeting Assessment in vitro and in vivo
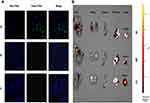
KLA induces cell apoptosis by disrupting mitochondria, but has poor permeability to cell membranes and is difficult to enter cells.16 Fluorescence microscopy imaging results showed that KLA conjugated to the selected peptide HMNPWSD could enter the cells, resulting in a strong signal accumulation inside the cells (Figure 2A). In addition, an evident signal enrichment was observed in the tumor of nude mice bearing PANC-1 tumor (Figure 2B), indicating that the fusion peptide HMNPWSD-KLA had strong targeting ability on PANC-1 cells in vitro and in vivo.
Toxicity of Fusion Peptide on PANC-1 Cells in vitro
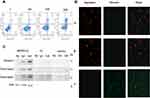
To assess the toxic effects of the fusion peptide on PANC-1 cells, PANC-1 and H1299 cells were subjected to MTT assay, with H1299 used as control cells. The MTT results showed that PANC-1 cell viability was inhibited in a dose-dependent manner by the treatment with the fusion peptide, while the H1299 cell viability was not affected when the concentration of the fusion peptide was less than 60 μM (Figure 3A and B). Thus, this was an additional proof further proving that the fusion peptide had a targeted inhibitory effect on PANC-1 cells.
Effect of Fusion Peptide on Cells and Mitochondria
Annexin V-FITC (green) and PI (red) can be used together to distinguish living cells (FITC/PI negative), early apoptotic cells (FITC positive, PI negative), late apoptotic cells or dead cells (FITC/PI positive). Flow cytometry analysis demonstrated that the fusion peptide was able to induce apoptosis in PANC-1 cells in a dose-dependent manner (Figure 4A). Mitochondrial membrane potential declined in the early stage of apoptosis, JC-1 probe was used to detect whether the apoptosis induced by the fusion peptide was related to mitochondrial damage. As shown in Figure 4B, the PANC-1 cells incubated with the fusion peptide emitted a green fluorescence, while the control group was characterized by a red fluorescence, indicating that the fusion peptide caused damage to the mitochondria of PANC-1 cells. Furthermore, the apoptotic proteins cytochrome c, cleaved caspase-3 and cleaved caspase-9 were activated in the PANC-1 cells after the treatment with the fusion peptide HMNPWSD-KLA (Figure 4C).
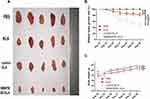
Effect of Fusion Peptides on Tumor Growth in vivo
The PANC-1 tumor model was treated with different peptides for 6 weeks, and the fusion peptide HMNPWSD-KLA showed a significant tumor growth inhibition compared with the effect of the other peptides (Figure 5A and B). The relative tumor growth rate (T/C) was used to evaluate the inhibitory effect of the peptide on tumor growth, and the results showed that T/C <60 is effective. Moreover, the weight of nude mice did not change significantly among the different group during the entire treatment period (Figure 5C).
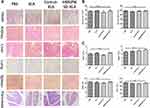
Adverse Effects of Fusion Peptides on the System
HE staining of vital organs (Figure 6A), liver enzyme index (Figure 6B) and hematological parameters (Figure 6C) were evaluated to establish the effects of the fusion peptide on normal tissues in nude mice. The fusion peptide did not have apparent adverse effects compared with the control group.
Discussion
Even if surgery can achieve R0 resection, some patients experience tumor recurrence and distant metastasis, affecting the postoperative survival rate.17 Chemotherapy plays an important role in increasing life expectancy of pancreatic cancer patients. However, due to the lack of specificity of chemotherapeutic drugs, there are many limitations: (1) they also kill healthy cells undergoing rapid proliferation resulting in toxic effects. (2) The development of the resistance to chemotherapeutic drugs by tumor cells.18–20 KLA has many advantages as a therapeutic peptide. First, KLA has low cytotoxicity due to its inability to effectively permeate the eukaryotic plasma membrane in mammalian cells.21 Second, KLA has a lower tendency to develop resistance due to its structure of only 14 amino acids, thus, with low immunogenicity.22 Third, it is easy to synthesize.
KLA requires the assistance of other cell-penetrating peptides for an effective crossing of the plasma membrane to induce apoptosis.23 In order to solve this problem, the peptide HMNPWSD that targeted pancreatic cancer cells and entered the cytoplasm was screened using phage display technology. We obtained 6 peptides by phage display technology at first. We conducted ELISA to evaluate the affinity of the 6 peptides for PANC-1 cells. According to the results (Figure 1A), the affinity of HMNPWSD and TAPHRSL for PANC-1 cells was much higher than that of the other four peptides. However, many peptides screened by phage display technology are non-specific binding to target cells or with weak affinity which cannot be completely eliminated in the biopanning part. That is why TAPHRSL has a high affinity to PANC-1 cells in ELISA but has weak signal in fluorescence staining experiment (Figure 1B). First, the targeting and position of the peptide was evaluated. After the peptide HMNPWSD was conjugated to KLA, KLA could enter the PANC-1 cells both in vitro and in vivo with the facilitate of HMNPWSD.
Subsequently, a series of experiments was performed on the biological effects of the fusion peptide. First, the fusion peptide showed a specific cytotoxicity on PANC-1 cells and had no effect on H1299 cell. This result also confirmed the specificity of HMNPWSD to PANC-1 cells from the side and the reduced toxic effects on normal tissues. Second, the experiments on the mechanism of KLA cytotoxicity confirmed that the fusion peptide induced PANC-1 apoptosis by disrupting mitochondria. As expected, animal experiment results showed that the fusion peptide significantly inhibited the growth of PANC-1 tumors without exerting any adverse effect on the system.
Initially, we intended to use targeting peptides for imaging diagnosis of early pancreatic cancer. We found that when we injected the fusion peptide through the tail vein to tumor-bearing mice with smaller tumors (<0.5 cm3), the fluorescence did not aggregate significantly as in larger tumors and the fluorescence intensity was relatively weak. It may be related to the relatively small number of PANC-1 cells, which is difficult to cause a large amount of aggregation of fluorescence. It is suggested that we need to modify the peptide and repeat the experiment with relatively stronger fluorescent dyes.
Conclusion
In conclusion, a peptide targeting pancreatic cancer cells was screened and conjugated with KLA, which specifically inhibited the growth of pancreatic cancer cells with minimal adverse effects. Thus, this approach possesses a great potential for targeted diagnosis and treatment of pancreatic cancer.
Author Contributions
Lei Zheng and Bo Zhang contributed equally to this work. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zeng H, Chen W, Zheng R, et al. Changing cancer survival in China during 2003–15: a pooled analysis of 17 population-based cancer registries. Lancet Glob Health. 2018;6(5):e555–e567. doi:10.1016/S2214-109X(18)30127-X
2. Chin V, Nagrial A, Sjoquist K, et al. Chemotherapy and radiotherapy for advanced pancreatic cancer. Cochrane Database Syst Rev. 2018;3:CD011044.
3. Kamisawa T, Wood LD, Itoi T, et al. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
4. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. Ca cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
5. Hanada K, Okazaki A, Hirano N, et al. Diagnostic strategies for early pancreatic cancer. J Gastroenterol. 2015;50(50):147–154. doi:10.1007/s00535-014-1026-z
6. Zhao YP. The present situation and the future of diagnosis and treatment of pancreatic cancer. Zhonghua Waike Zazhi. 2014;52:641–643.
7. Huang PH, Lu PJ, Ding LY, et al. TGFβ promotes mesenchymal phenotype of pancreatic cancer cells, in part, through epigenetic activation of VAV1. Oncogene. 2017;36(16):2202–2214. doi:10.1038/onc.2016.378
8. Huang L, Jansen L, balavarca Y, et al. Resection of pancreatic cancer in Europe and USA: an international largescale study highlighting large variations. Gut. 2017;265(4):800–805.
9. Liu S, Chen S, Zeng J. TGF-β signaling; a complex role in tumorigenesis (review). Mol Med Rep. 2018;17(1):699–704. doi:10.3892/mmr.2017.7970
10. Ueberberg S, Meier JJ, Waengler C, et al. Generation of novel single-chain antibodies by phage-display technology to direct imaging agents highly selective to pancreatic beta- or alpha cells in vivo. Diabetes. 2009;58(10):2324–2334. doi:10.2337/db09-0658
11. Rahbarnia L, Farajnia S, Babaei H, et al. Evolution of phage display technology: from discovery to application. J Drug Target. 2017;25:216–224. doi:10.1080/1061186X.2016.1258570
12. Xiao YF, Jie MM, Li BS, et al. Peptide-based treatment: a promising cancer therapy. J Immunol Res. 2015;2015:1–13. doi:10.1155/2015/761820
13. Marqus S, Pirogova E, Piva TJ. Evaluation of the use of therapeutic peptides for cancer treatment. J Biomed Sci. 2017;24:21. doi:10.1186/s12929-017-0328-x
14. Ellerby HM, Arap W, Ellerby LM, et al. Anti-cancer activity of targeted pro-apoptotic peptides. Nat Med. 1999;5(9):1032–1038. doi:10.1038/12469
15. Chen WH, Xu X-D, Luo G-F, et al. Dual-targeting pro-apoptotic peptide for programmed cancer cell death via specific mitochondria damage. Sci Rep. 2013;3:3468. doi:10.1038/srep03468
16. Cieslewicz M, Tang J, Yu JL, et al. Targeted delivery of proapoptotic peptides to tumor associated macrophages improves survival. Proc Natl Acad Sci U S A. 2013;110:15919–15924. doi:10.1073/pnas.1312197110
17. Yan JF, Xu XW, Jin WW, et al. Laparoscopic spleen-preserving distal pancreatectomy for pancreatic neoplasms: a retrospective study. World J Gastroenterol. 2014;20:13966–13972. doi:10.3748/wjg.v20.i38.13966
18. Chism DD, De Silva D, Whang YE. Mechanisms of acquired resistance to androgen receptor targeting drugs in castration-resistant prostate cancer. Expert Rev Anticancer Ther. 2014;14:1369–1378. doi:10.1586/14737140.2014.928594
19. Tamburrino A, Piro G, Carbone C, Tortora G, Melisi D. Mechanisms of resistance to chemotherapeutic and anti-angiogenic drugs as novel targets for pancreatic cancer therapy. Front Pharmacol. 2013;4:56. doi:10.3389/fphar.2013.00056
20. Chatterjee S, Damle SG, Sharma AK. Mechanisms of resistance against cancer therapeutic drugs. Curr Pharm Biotechnol. 2014;15:1105–1112. doi:10.2174/1389201015666141126123952
21. Hyun S, Lee S, Kim S, Jang S, Yu J, Lee Y. Apoptosis inducing, conformationally constrained, dimeric peptide analogs of KLA with submicromolar cell penetrating abilities. Biomacromolecules. 2014;15(10):3746–3752. doi:10.1021/bm501026e
22. Shagaghi N, Palombo EA, Clayton AH, Bhave M. Archetypal tryptophan-rich antimicrobial peptides: properties and applications. World J Microbiol Biotechnol. 2016;32:31. doi:10.1007/s11274-015-1986-z
23. Law B, Quinti L, Choi Y, Weissleder R, Tung C-H. A mitochondrial targeted fusion peptide exhibits remarkable cytotoxicity. Mol Cancer Ther. 2006;5(8):1944–1949. doi:10.1158/1535-7163.MCT-05-0509
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.