Back to Journals » OncoTargets and Therapy » Volume 9
A genetic variant in large tumor suppressor kinase 2 of Hippo signaling pathway contributes to prognosis of hepatocellular carcinoma
Authors Shen L, Wen J, Zhao T , Hu Z , Song C, Gu D, He M, Lee N, Xu Z, Chen J
Received 17 November 2015
Accepted for publication 10 January 2016
Published 4 April 2016 Volume 2016:9 Pages 1945—1951
DOI https://doi.org/10.2147/OTT.S100699
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr William C. Cho
Lili Shen,1,2,* Juan Wen,3,4,* Tingting Zhao,1 Zhibin Hu,4,5 Ci Song,4 Dongying Gu,1 Mingliang He,6 Nikki P Lee,7 Zhi Xu,1 Jinfei Chen1
1Department of Oncology, Nanjing First Hospital, Nanjing Medical University, Nanjing, 2Department of Oncology, Haimen People’s Hospital, Haimen, 3Nanjing Maternity and Child Health Care Institute, Nanjing Maternity and Child Health Care Hospital Affiliated with Nanjing Medical University, Nanjing, 4Department of Epidemiology and Biostatistics, Jiangsu Key Laboratory of Cancer Biomarkers, Prevention and Treatment, Cancer Center, School of Public Health, Nanjing Medical University, Nanjing, 5State Key Laboratory of Reproductive Medicine, Nanjing Medical University, Nanjing, 6Department of Biomedical Sciences, City University of Hong Kong, Hong Kong, 7Department of Surgery, The University of Hong Kong, Hong Kong, People’s Republic of China
*These authors contributed equally to this work
Abstract: The Hippo pathway plays an important role in the development of hepatocellular carcinoma (HCC). The present study aimed at exploring the genetic variants of Hippo pathway-related genes and their association with HCC prognosis. A total of 331 HCC patients who tested positive for hepatitis B surface antigen were recruited in this study. None of the patients had prior surgical treatment. Twelve potentially functional single-nucleotide polymorphisms (rs7317471 and rs9509492 in LATS2; rs4810446, rs2267853, rs8000, and rs6073627 in MST1; rs10955176 in MST2; and rs16861979, rs2043550, rs16861985, rs1055153, and rs7630434 in TAZ) in the Hippo pathway were genotyped from patients’ peripheral leukocytes using the Sequenom MassARRAY iPLEX platform. Cox proportional hazard models and log-rank test were used for the survival analyses. LATS2 rs7317471 C>T polymorphism was significantly associated with decreased risk of death in HCC using the dominant model (adjusted hazard ratio [HR] =0.63, 95% confidence interval [CI] =0.46–0.87, P=0.004). Furthermore, using stratified analysis, LATS2 rs7317471 CT/TT genotypes were found to be significantly associated with decreased risk of death in patients who were below 53 years of age (adjusted HR =0.50), females (adjusted HR =0.60), smokers (adjusted HR =0.56), drinkers (adjusted HR =0.58), have Barcelona clinic liver cancer stage B (adjusted HR =0.62), and received no prior chemotherapy or transcatheter hepatic arterial chemoembolization (adjusted HR =0.48). Our results suggested that LATS2 rs7317471 could be used as a potential biomarker for the prediction of HCC prognosis.
Keywords: hepatocellular carcinoma, LATS2, prognosis, retrospective study, single-nucleotide polymorphism
Introduction
Liver cancer is a prevalent gastrointestinal tract cancer in China, with a total of 355,595 newly diagnosed cases and 322,417 deaths reported in 2011.1 The most common histological type of liver cancer is hepatocellular carcinoma (HCC). Even with the implementation of new treatment strategies including liver transplantation, surgical resection, chemotherapy, and molecular targeting therapy, the prognosis for HCC patients remains poor.2 To facilitate better management of HCC patients, it is crucial to identify specific biomarkers for prognosis prediction.
Hippo pathway is composed of a network of molecules that regulate HCC cell proliferation, apoptosis, migration, and differentiation.3 The core of the mammalian Hippo pathway is a kinase cassette comprising mammalian STE20-like protein kinase 1/2 (MST1/2), salvador family WW domain containing protein 1 (SAV1), large tumor suppressor kinase 1/2 (LATS1/2), and MOB kinase activator 1 (MOB1). Upon activation of the Hippo pathway, MST1/2-SAV1 phosphorylates and activates LATS1/2. The subsequent binding of LATS1/2 with MOB1 will then phosphorylate the Yes-associated protein (YAP) and tafazzin (TAZ), leading to their cytoplasmic retention. In this way, excessive cell proliferation and tumorigenesis will be inhibited.4,5 When components of the core kinase cassette are dysregulated, hypophosphorylated YAP will be translocated into the nucleus and will directly interact with the transcription factors, which will then induce a number of downstream gene expressions involving cell proliferation and cell death. As a result of excessive cell proliferation, aberrant YAP activation is associated with the development of multiple tumors including those of the liver,6 lung,7 ovary,8 colon–rectum,9 pancreas,10 and prostate.11 Meanwhile, Hippo pathway may also be subject to regulation by any genetic alterations that manipulate the expression and function of its core components. An inactive cleaved form of MST1 was found in HCC tumors and correlated with low levels of phosphorylated YAP.12 The single-nucleotide polymorphism (SNP) of YAP rs1820453 A>C may affect its expression levels and is associated with increased risk of breast cancer in the Chinese population.13 This polymorphism was also found to be associated with reduced YAP expression and prognosis of small-cell lung cancer patients treated with platinum-based chemotherapy.14
To systematically investigate the clinical significance of genetic variations of Hippo pathway components in HCC, we performed SNP genotyping on 12 potentially functional genetic variants of the Hippo pathway in 331 HCC patients. Statistical analyses were performed to examine the prognostic value of each genetic variant in HCC.
Materials and methods
Patients
A total of 331 HCC patients were recruited from Nantong Tumor Hospital (Nantong, People’s Republic of China) and the First Affiliated Hospital of Nanjing Medical University, (Nanjing, People’s Republic of China) from January 2006 to December 2010. Patients were diagnosed with HCC based on histopathological examination, or the measurement of serological α-fetoprotein level (>400 ng/mL) and imaging examination by magnetic resonance imaging and/or computerized tomography, as described previously.15–17 In brief, patients enrolled in this study had either intermediate or advanced tumor stage according to Barcelona Clinic Liver Cancer (BCLC) staging system and tested positive for hepatitis B surface antigen.18 Patients were followed up every 3 months from the date of enrollment until death or to the last follow-up date (January 14, 2013). Individuals who smoked 1 cigarette per day for over 1 year were defined as smokers, and those who consumed one or more alcohol drinks a week for over 6 months were categorized as alcohol drinkers. None of them had received surgery. The study was approved by the institutional review board of Nanjing Medical University (Nanjing, People’s Republic of China). Signed informed consents were collected from enrolled patients for the use of clinical specimens in medical research.
Serological tests
Surface antigen of hepatitis B virus (HBV), antibody against HBV surface antigen, antibody against HBV core antigen, and antibody against hepatitis C virus were detected by enzyme-linked immunosorbent assays (Kehua Bio-Engineering Co., Ltd., Shanghai, People’s Republic of China), following the manufacturer’s instructions.19
Selection and genotyping of SNPs
Common SNPs associated with core components (of LATS1/2, MST1/2, SAV1, MOB1, YAP, and TAZ) of Hippo signaling were included for initial analysis. The selected SNPs had minor allele frequency larger than 0.05 in Chinese/Asians and were located at the 5′ flanking regions, 5′-untranslated regions (5′-UTRs), coding regions, or 3′-UTRs according to NCBI dbSNP data (last search date: November 2014). We also included SNPs with biological significance or those that were associated with gene expression and/or cancer risk/survival according to the published literature. If SNPs are in high linkage disequilibrium (r2>0.8), we would genotype only one SNP. As a result, 12 SNPs (rs7317471 and rs9509492 in LATS2; rs4810446, rs2267853, rs8000, and rs6073627 in MST1; rs10955176 in MST2; and rs16861979, rs2043550, rs16861985, rs1055153, and rs7630434 in TAZ) were selected for genotyping (Table 1).
Genomic DNA was extracted from a leukocyte pellet by traditional proteinase K digestion, phenol–chloroform extraction, and ethanol precipitation. All SNPs were genotyped using the Sequenom MassARRAY iPLEX platform (Sequenom, Inc., San Diego, CA, USA). All primers used are listed in Table S1. Two blank (water) controls in each 384-well plate were performed for quality control, and more than 5% samples were randomly selected and the processes repeated, yielding a 100% concordance. The success rates of genotyping all 12 SNPs were above 95%.
Statistical analyses
Survival time was calculated from the date of HCC diagnosis to the date of last follow-up or until death. Hardy–Weinberg equilibrium was assessed within patients by using a goodness-of-fit χ2 test. Median overall survival time (MOST) was calculated, and mean survival time was presented when the MOST could not be calculated. Log-rank test was used to compare the survival time in different subgroups categorized by patient characteristics, clinical features, and genotypes. Univariate and multivariate Cox proportional hazard regression analyses were performed to estimate the crude or adjusted hazard ratio (HR) and their 95% confidence intervals (CIs), with adjustment of age, sex, smoking status, drinking status, BCLC tumor stage, and chemotherapy or intervention status. The significance level was set at P<0.05, and P-values were two-sided. Analyses were carried out using the Statistical Analysis System software (version 9.1.3; SAS Institute, Cary, NC, USA).
Results
Clinical features of the studied patient cohort
The demographic characteristics and clinical features of the 331 HCC patients were described previously.17 In this cohort, 284 (85.8%) patients were male and 47 (14.2%) were female. Three hundred and four patients (91.8%) had BCLC stage B disease, and 27 (8.2%) patients had BCLC stage C disease. Chemotherapy or interventional therapy was given to 91 patients (27.5%). The MOST of the cohort was 14.5 months. Patients who were defined as alcohol drinkers had a shorter median overall survival time (MOST, 12.6 months) than nondrinkers (HR =1.43, 95% CI =1.11–1.84, P=0.006). Compared to patients without chemotherapy or intervention (transcatheter arterial chemoembolization [TACE]) therapy (MOST =3.4 months), patients treated with chemotherapy or intervention therapy (MOST =16.8 months) had a decreased risk of death by 61% (HR =0.39, 95% CI =0.29–0.51, P<0.001).
SNPs associated with patient survival in HCC
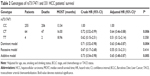
Kaplan–Meier and log-rank tests were used to examine the associations between the 12 studied SNPs and HCC survival by using different genetic models. All P-values of Hardy–Weinberg equilibrium were greater than 0.05. As shown in Table 1, only LATS2 rs7317471 C>T polymorphism was significantly correlated with the overall survival time of HCC patients using the dominant model (Figure 1), while the rest of the SNPs were not found to have significant correlations with the overall survival time. After adjusting for age, sex, smoking and drinking status, BCLC tumor stage, and chemotherapy or TACE status, LATS2 rs7317471 CT/TT genotypes remained significantly associated with favorable survival outcome (adjusted HR =0.63, 95% CI =0.46–0.87, P=0.004; Table 2) but not others.
  | Figure 1 Kaplan–Meier plots of survival by LATS2 rs7317471 genotypes in HCC patients’ survival (log-rank P=0.034 for CT/TT vs CC) in a dominant model. |
To better assess the correlation between LATS2 rs7317471 polymorphism and HCC survival, stratified analysis was performed on age, sex, smoking and drinking status, BCLC tumor stage, and chemotherapy or TACE status. As shown in Table 3, HCC patients who were younger than 53 years of age and had LATS2 rs7317471 CT/TT genotypes demonstrated longer overall survival (adjusted HR =0.50, 95% CI =0.31–0.81). Other stratified factors such as being female (adjusted HR =0.60, 95% CI =0.42–0.85) or smokers (adjusted HR =0.56, 95% CI =0.37–0.83) or drinkers (adjusted HR =0.58, 95% CI =0.38–0.87) or having BCLC stage B disease (adjusted HR =0.62, 95% CI =0.44–0.86) and having not received prior chemotherapy or TACE treatment (adjusted HR =0.48, 95% CI =0.25–0.91), all demonstrated similar outcomes.
Multivariate Cox regression analysis on HCC survival
Multivariate Cox regression analysis was performed to analyze the effects of clinicopathological data, treatment, and LATS2 rs7317471 on HCC survival. In addition to age (P=0.005), chemotherapy or TACE treatment (P<0.001), drinking status (P=0.004), and LATS2 rs7317471 were found to be independent prognostic markers for the overall survival of HCC patients (HR =0.62, 95% CI =0.45–0.85, P=0.003; Table 4).
Discussion
In this study, we investigated the correlations between the functional polymorphisms of the core components in Hippo pathway with overall survival in 331 Chinese HCC patients. Among the 12 selected SNPs, eight SNPs were located in introns (rs4810446, rs2267853, rs8000, and rs6073627 in MST1; rs16861979, rs2043550, and rs16861985 in TAZ; and rs9509492 in LATS2), two of them (rs10955176 in MST2 and rs7630434 in TAZ) were located at the 3′-UTR, rs1055153 was located at the 5′ end of TAZ, and rs7317471 was located in the coding region of LATS2. In this cohort, LATS2 rs7317471was found to be a prognostic biomarker for HCC survival in Chinese patients.
In our research, LATS2 rs7317471 C>T polymorphism correlated significantly with survival of HCC patients. Compared to CC genotype, HCC patients with rs7317471 CT/TT genotypes tended to have a lower risk of death. This association was more prominent in the subgroups of patients who were of younger age, were females, smokers, or drinkers, had BCLC tumor stage B, and did not receive chemotherapy or TACE treatments. In addition, multivariate Cox regression analysis showed that LATS2 rs7317471 polymorphism was an independent biomarker for survival of HCC patients.
LATS2 is a serine/threonine kinase located on 13q12.11.20–23 LATS2 acts as a putative tumor suppressor that controls G1/S and G2/M transition of the cell cycle.24,25 Besides, it can also suppress antiapoptotic proteins Bcl-2 and Bcl-XL to induce apoptosis as well as interacting with other cell division regulators, such as p53, to maintain mitotic fidelity and genetic stability.26,27 Hence, LATS2 malfunction will lead to aberrant cell proliferation, which is one of the hallmarks in cancer development and progression. LATS2 expression is associated with several types of tumors including breast cancer,13 prostate cancer,28 esophageal cancer, and colorectal cancer.29 Our study showed that LATS2 rs7317471 C>T polymorphism was related with decreased risk of death in Chinese HCC patients, and it might also be associated with the aberrant expression or malfunction of LATS2 caused by this base change. Further research to investigate the function of this LATS2 variant is needed.
Conclusion
To our knowledge, this is the first study to investigate the association between SNPs of the core Hippo pathway components with HCC survival. Among the 12 SNPs, LATS2 rs7317471 polymorphism decreased risk of death of HCC patients younger than 53 years of age, females, smokers, drinkers, those with BCLC stage B, and those who had not received chemotherapy or TACE. However, there are several limitations in our study. The results are based on a relatively small cohort of only 331 Chinese HBV-related HCC patients; further validation will be needed in a larger cohort. Meanwhile, the function of LATS2 rs7317471 has not been characterized. Investigating the consequence of this polymorphism on LATS2 expression and function as well as examining the correlations between LATS2 rs7317471 genotypes and the alterations of Hippo pathway components may help to reveal the functional role of this polymorphism. In summary, our results indicate that LATS2 rs7317471 could be a potential prognostic biomarker for HCC patients in China.
Disclosure
This study was supported by grants from the National Natural Science Foundation of China (81370057 to Zhi Xu and 81272469 to Jinfei Chen), the Key Project supported by Medical Science and Technology Development Foundation, Nanjing Department of Health (JQX12007 to Zhi Xu), the clinical special project for Natural Science Foundation of Jiangsu Province (BL2012016 to Jinfei Chen), and the grant from Nanjing 12th Five-Year Key Scientific Project of Medicine to Jinfei Chen. The authors report no conflicts of interest in this work.
References
Chen W, Zheng R, Zeng H, Zhang S, He J. Annual report on status of cancer in China, 2011. Chin J Cancer Res. 2015;27(1):2–12. | ||
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Harvey KF, Zhang X, Thomas DM. The hippo pathway and human cancer. Nat Rev Cancer. 2013;13(4):246–257. | ||
Hong W, Guan KL. The yap and taz transcription co-activators: key downstream effectors of the mammalian hippo pathway. Semin Cell Dev Biol. 2012;23(7):785–793. | ||
Zhou D, Zhang Y, Wu H, et al. Mst1 and mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A. 2011;108(49):1312–1320. | ||
Perra A, Kowalik MA, Ghiso E, et al. YAP activation is an early event and a potential therapeutic target in liver cancer development. J Hepatol. 2014;61(5):1088–1096. | ||
Lau AN, Curtis SJ, Fillmore CM, et al. Tumor-propagating cells and Yap/Taz activity contribute to lung tumor progression and metastasis. EMBO J. 2014;33(5):468–481. | ||
Hall CA, Wang R, Miao J, et al. Hippo pathway effector Yap is an ovarian cancer oncogene. Cancer Res. 2010;70(21):8517–8525. | ||
Avruch J, Zhou D, Bardeesy N. YAP oncogene overexpression supercharges colon cancer proliferation. Cell Cycle. 2012;11(6):1090–1096. | ||
Zhang W, Nandakumar N, Shi Y, et al. Downstream of mutant KRAS, the transcription regulator YAP is essential for neoplastic progression to pancreatic ductal adenocarcinoma. Sci Signal. 2014;7(324):ra42. | ||
Zhang L, Yang S, Chen X, et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol Cell Biol. 2015;35(8):1350–1362. | ||
Zhou D, Conrad C, Xia F, et al. MST1 and MST2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through in activation of the YAP1 oncogene. Cancer Cell. 2009;16(5):425–438. | ||
Chen W, Wang W, Zhu B, et al. A functional variant rs1820453 in YAP1 and breast cancer risk in Chinese population. PLoS One. 2013;8(11):e79056. | ||
Wu C, Xu B, Yuan P, et al. Genome-wide interrogation identifies YAP1 variants associated with survival of small-cell lung cancer patients. Cancer Res. 2010;70(23):9721–9729. | ||
Pan Y, Sun C, Huang M, et al. A genetic variant in pseudogene e2f3p1 contributes to prognosis of hepatocellular carcinoma. J Biomed Res. 2014;28(3):194–200. | ||
Qi F, Huang M, Pan Y, et al. A genetic variant in the promoter region of mir-106b-25 cluster predict clinical outcome of hbv-related hepatocellular carcinoma in Chinese. PLoS One. 2014;9(1):e85394. | ||
Xie K, Liu J, Zhu L, et al. A potentially functional polymorphism in the promoter region of let-7 family is associated with survival of hepatocellular carcinoma. Cancer Epidemiol. 2013;37(6):998–1002. | ||
Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. | ||
Hu L, Zhai X, Liu J, et al. Genetic variants in human leukocyte antigen/DP-DQ influence both hepatitis B virus clearance and hepatocellular carcinoma development. Hepatology. 2012;55(5):1426–1431. | ||
Hori T, Takaori-Kondo A, Kamikubo Y, Uchiyama T. Molecular cloning of a novel human protein kinase, kpm, that is homologous to warts/lats, a Drosophila tumor suppressor. Oncogene. 2000;19(27):3101–3109. | ||
Dong J, Feldmann G, Huang J, et al. Elucidation of a universal size-control mechanism in drosophila and mammals. Cell. 2007;130(6):1120–1133. | ||
Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9(5):534–546. | ||
Xu T, Wang W, Zhang S, Stewart RA, Yu W. Identifying tumor suppressors in genetic mosaics: the drosophila lats gene encodes a putative protein kinase. Development. 1995;121(4):1053–1063. | ||
Kamikubo Y, Takaori-Kondo A, Uchiyama T, Hori T. Inhibition of cell growth by conditional expression of kpm, a human homologue of drosophila warts/lats tumor suppressor. J Biol Chem. 2003;278(20):17609–17614. | ||
Li Y, Pei J, Xia H, Ke H, Wang H, Tao W. Lats2, a putative tumor suppressor, inhibits g1/s transition. Oncogene. 2003;22(28):4398–4405. | ||
Ke H, Pei J, Ni Z, et al. Putative tumor suppressor lats2 induces apoptosis through downregulation of bcl-2 and bcl-x(l). Exp Cell Res. 2004;298(2):329–338. | ||
Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20(19):2687–2700. | ||
Powzaniuk M, McElwee-Witmer S, Vogel RL, et al. The lats2/kpm tumor suppressor is a negative regulator of the androgen receptor. Mol Endocrinol. 2004;18(8):2011–2023. | ||
Lee KH, Goan YG, Hsiao M, et al. Microrna-373 (mir-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (lats2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315(15):2529–2538. |
Supplementary material
  | Table S1 Primers and probes for Sequenom allelic discrimination |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.