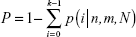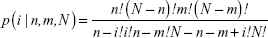Back to Journals » OncoTargets and Therapy » Volume 10
A comprehensive insight into the clinicopathologic significance of miR-144-3p in hepatocellular carcinoma
Authors Liang HW, Ye ZH, Yin SY, Mo WJ, Wang HL, Zhao JC, Liang GM, Feng ZB, Chen G , Luo DZ
Received 28 March 2017
Accepted for publication 10 June 2017
Published 11 July 2017 Volume 2017:10 Pages 3405—3419
DOI https://doi.org/10.2147/OTT.S138143
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Samir Farghaly
Hai-wei Liang,* Zhi-hua Ye,* Shu-ya Yin, Wei-jia Mo, Han-lin Wang, Jin-che Zhao, Guo-mei Liang, Zhen-bo Feng, Gang Chen, Dian-zhong Luo
Department of Pathology, First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, China
*These authors contributed equally to this work
Background: Studies which focused on the character of miR-144-3p in hepatocellular carcinoma (HCC) are limited. This study aimed to explore the expression, clinical significance and the potential targets of miR-144-3p in HCC.
Methods: The Cancer Genome Atlas (TCGA) and a cohort of 95 cases of HCC were applied to investigate aberrant miR-144-3p expression in HCC. A meta-analysis was performed to accumulate data on miR-144-3p expression in HCC based on TCGA, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Gene Expression Omnibus (GEO). Additionally, the potential regulatory mechanisms of miR-144-3p in HCC were explored by bioinformatics.
Results: MiR-144-3p expression was downregulated distinctly in HCC compared to para-HCC tissue both in TCGA data (8.9139±1.5986 vs 10.7721±0.9156, P<0.001) and in our qRT-PCR validation (1.3208±0.7594 vs 2.6200±0.9263, P<0.001). The meta-analysis based on TCGA, qRT-PCR and GEO data confirmed a consistent result (standard mean difference =-0.854, 95% CI: -1.224 to -0.484, P<0.001). The receiver operating characteristic curve of miR-144-3p gained a significant diagnostic value both in TCGA data (area under the curve [AUC] =0.852, 95% CI: 0.810 to 0.894, P<0.001) and in qRT-PCR validation (AUC =0.867, 95% CI: 0.817 to 0.916, P<0.001), especially in alpha-fetoprotein–negative HCC patients (AUC =0.900, 95% CI: 0.839 to 0.960, P<0.001). Furthermore, we identified 119 potential targets of miR-144-3p in HCC by bioinformatics. Gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses revealed that several significant biologic functions and pathways correlated with the pathogenesis of HCC, including the p53 signaling pathway.
Conclusion: MiR-144-3p may function as a cancer suppressor microRNA, which is essential for HCC progression through the regulation of various signaling pathways. Thus, interactions with miR-144-3p may provide a novel treatment strategy for HCC in the future.
Keywords: miR-144-3p, hepatocellular carcinoma, TCGA, qRT-PCR, GEO, gene functional enrichment analysis
Introduction
Hepatocellular carcinoma (HCC) is a common malignancy with a high incidence and unsatisfactory prognosis worldwide.1,2 In the past few decades, surgery was considered the first-line treatment of HCC.3,4 However, most HCC patients were diagnosed in advanced stage and, therefore, lost the chance for surgery.5–7 Up to now, it still remains a challenge to improve the survival of HCC patients.8 Despite great progress in the treatment of HCC, including interventional therapy and molecular target treatment, only one molecular targeted agent (sorafenib) has been demonstrated to improve the survival of HCC.9–13 Consequently, it is worthwhile to identify novel molecular targets to facilitate early diagnosis and targeted therapy of HCC to achieve a better survival of HCC patients.
MicroRNAs (miRNAs) are a family of noncoding RNAs which can take part in cell apoptosis and proliferation.14–19 Recently, it has been reported that miRNAs can play an indispensable role in the pathogenesis of cancers through the regulation of oncogene and anti-oncogene.20–28 There have been three published articles that investigated miR-144-3p level in HCC tissue with small sample size, and all of them showed downregulation of miR-144-3p expression in HCC.29–31 However, the limited sample size of the three individual studies (23 pairs, 33 pairs and 100 pairs, respectively) may lead to false-positive results and a weak conclusion. Hence, attempts to verify the altered miR-144-3p expression in HCC are necessary.
Up to now, several studies have investigated the clinical impact of miR-144-3p in several cancers, and all demonstrated the important role of miR-144-3p in carcinogenesis. In lung cancer, miR-144-3p could inhibit cell proliferation and induce apoptosis and autophagy. In bladder cancer, Guo et al64 clarified that miR-144-3p could regulate Wnt signaling pathway to promote cancer cell proliferation. In prostate cancer, Gu et al65 clarified that miR-144-3p was associated with radiosensitivity. However, the investigation into the role of miR-144-3p in HCC is rare. Thus, in this study, we focused on the role of miR-144-3p in HCC.
In this study, we investigated the miR-144-3p level in HCC from three sources (The Cancer Genome Atlas [TCGA] database, quantitative reverse transcription-polymerase chain reaction [qRT-PCR] and Gene Expression Omnibus [GEO] database) and combined them by a meta-analysis. Afterward, the clinical significance of miR-144-3p in the diagnosis and prognosis of HCC was evaluated. Moreover, the underlying mechanisms of the pathogenesis of HCC were analyzed through identifying the potential target genes of miR-144-3p and performing a gene functional enrichment analysis.
Materials and methods
Patients and samples
We collected 95 HCC samples with matched adjacent nontumor tissues from 75 males and 20 females (mean age: 50 years) at the First Affiliated Hospital of Guangxi Medical University between March 2010 and March 2012. The patients in this cohort had not received any treatment before the surgery. Two pathologists performed the pathologic diagnoses independently. The study was approved by the ethics committee of the First Affiliated Hospital of Guangxi Medical University, Nanning, China, and all patients signed the written informed consent for our study.
MiR-144-3p expression in HCC in TCGA
We mined TCGA database (https://cancergenome.nih.gov/) to explore miR-144-3p expression in HCC tissues. The level three data of the miRNA profile of HCC with the corresponding clinical data were evaluated. Next, miR-144 expression was selected from the miRNA profile. Subsequently, we conducted the statistical analyses to assess miR-144 expression in HCC and the associations between miR-144 level and the clinicopathologic features.
Quantitative reverse transcription-polymerase chain reaction
The qRT-PCR procedure has been reported in our previous studies.20,22,32–39 A combination of RUN6B and RUN48, which has been considered a stable internal control in our previous studies, was utilized as a reference for the detection of miR-144-3p levels in this study.20,35,40 The RNU6B, RNU48 and miR-144-3p primers were involved in the TaqMan MicroRNA Assays (4427975; Thermo Fisher Scientific, Waltham, MA, USA), and their sequences were as follows: miR-144-3p, UACAGUAUAGAUGAUGUACU; RNU6B, CGCAAGGAUGACACGCAAAUUCGUGAAGCGUUCCAUAUUUUU; RNU48, GAUGACCCCAGGUAACUCUGAGUGUGUCGCUGAUGCCAUCACCGCAGCGCUCUGACC. The miR-144-3p level was computed by the formula 2−Δcq.20,22,32–39
MiR-144-3p expression in HCC in the GEO
We searched for miR-144-3p expression in HCC in GEO database with the following key words: 1) (hepatocellular OR liver OR hepatic OR HCC), 2) malignan* OR cancer OR tumor OR tumour OR neoplas* OR carcinoma and 3) microRNA OR miRNA OR miR. The microarrays, which examined miR-144-3p expression in HCC tissues and noncancerous tissues, were included. The microarrays with poor quality were excluded. The microarray with poor quality was the one that 1) did not provide useful data for meta-analysis, 2) did not have control group and 3) only detected one sample of HCC. Finally, the included GEO dataset was GSE41874, GSE40744, GSE21362, GSE22058, GSE12717 and GSE57555.
Meta-analysis
Stata 12.0 software was used for the meta-analysis. The meta-analysis combined three sources (TCGA data, qRT-PCR data and GEO data) of the expression of miR-144-3p in HCC. Standard mean difference (SMD) with 95% CI was utilized to assess the pooled data in the meta-analysis. The Q test (chi-squared test) and I2 were calculated to judge the heterogeneity among the eligible studies. A fixed-effects model was performed if no obvious heterogeneity existed among the pooled studies (I2<50% and P>0.1) and a random-effect model was utilized when obvious heterogeneity was observed among the studies (I2>50% or P<0.1).41
MiRNA target prediction
Twelve databases (miRWalk, Microt4, miRanda, mirbridge, miRDB, miRMap, miRNAMap, Pictar2, PITA, RNAhybrid, Targetscan and miRWalk) were used for the target prediction of miR-144-3p,42 and the predicted target genes of miR-144-3p that were reported in at least four databases were selected. Three databases (miRTarbase, Tarbase and miRNet) were searched to obtain the validated targets of miR-144-3p.43–45 Next, we combined the two parts to identify the target genes of miR-144-3p and applied the target genes to further gene functional enrichment analysis.
Natural language processing (NLP) analysis of HCC-related studies
We retrieved the PubMed database with the following keywords: 1) hepatocellular OR liver OR hepatic OR HCC and 2) malignan* OR cancer OR tumor OR tumour OR neoplas* OR carcinoma. In the published studies, the HCC-related genes were identified through A Biomedical Named Entity Recognizer (http://pages.cs.wisc.edu/~bsettles/abner/).
As for the NLP analysis, we computed the frequency of all the genes in the included studies in PubMed database. A higher frequency of a certain gene represents a greater likelihood of the associations between HCC and the certain gene. The probability of a frequency greater than “k” co-citations was analyzed of random conditions via hypergeometric distribution. A P-value <0.01 showed statistically significance. The P-value of a certain HCC-related gene was calculated with the following formulas:
|
|
where N indicates all the eligible studies, m is the frequency of the related genes, n is the frequency of HCC and k is the co-occurrence of HCC and a certain gene.
Gene functional enrichment analysis by bioinformatics
Gene ontology (GO) analysis, including three sections (biologic process, molecular function and cellular component), was completed in DAVID database.46 Cytoscape software with BINGO contributed to the GO network. The protein-to-protein network was analyzed in STRING.47 The databases of the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Panther were also applied in the enrichment of the underlying biologic pathways of miR-144-3p targets.
Statistical analysis
We conducted all the statistical analyses in SPSS 22.0 software and used Student’s t-tests to analyze the quantitative miR-144-3p levels between two groups. One-way analysis of variance was utilized for three groups. Furthermore, we exploited receiver operating characteristic (ROC) curves and calculated the area under the curve (AUC) to estimate the diagnostic value of miR-144-3p for HCC. The standard to assess the efficacy for HCC diagnosis in ROC was as follows: low (AUC: 0.5–0.7), moderate (AUC: 0.7–0.9) and high (0.9–1.0). A P-value <0.05 indicated statistical difference.
Results
MiR-144-3p expression in HCC based on TCGA
Regarding TCGA data, decreased miR-144-3p levels were detected in 354 HCC tissues (8.9139±1.5986) compared to 50 adjacent normal liver tissues (10.7721±0.9156, P<0.001; Figure 1A). Not surprisingly, a consistent expression pattern was observed in 48 pairs of HCC and adjacent normal liver tissues (P<0.001; Figure 1B). The P-value of the diagnostic power in ROC curve was <0.001 (AUC =0.852, 95% CI: 0.810 to 0.894, P<0.001; Figure 2A). The results of the relationships between miR-144-3p and the clinicopathologic characteristics from TCGA data are summarized in Table 1. Low miR-144-3p expression was only detected in high pathologic stage rather than low pathologic stage. No significant correlation was observed between miR-144-3p and other clinicopathologic features. In the survival analyses by ProgMIR (http://xvm145.jefferson.edu/progmir/), no significant results were observed (hazard ratio =0.93, 95% CI: 0.83 to 1.04, P=0.204).
MiR-144-3p expression validation in HCC by qRT-PCR
In contrast to the adjacent normal liver tissue (2.6200±0.9263), miR-144-3p was significantly underexpressed in HCC tissue (1.3208±0.7594, P<0.001) with a significant ROC curve (AUC=0.867, 95% CI: 0.817 to 0.916, P<0.001), as shown in Table 2, Figures 2B and 3A.
All of the clinicopathologic factors are presented in Table 2. Regarding the associations between the expression of miR-144-3p and the clinical characteristics in HCC, we found that the downregulation of miR-144-3p was detected in HCC with metastasis (1.1120±0.5994, P=0.006; Figure 3B), advanced tumor node metastasis (TNM) stage (1.1596±0.5821, P=0.001; Figure 3C) and tumor capsular infiltration (1.1746±0.6991, P=0.047; Figure 3D), compared to the HCC without metastasis (1.5433±0.8504), early TNM stage (1.8559±1.0146) and without tumor capsular infiltration (1.4833±0.7977), respectively. The rest of the clinicopathologic parameters of HCC had no significant influence on the expression of miR-144-3p (Table 2).
Interestingly, we obtained two altered AUCs in alpha-fetoprotein (AFP)-positive HCC patients (AUC =0.827, 95% CI: 0.749 to 0.905, P<0.001; Figure 4A) and AFP-negative HCC patients (AUC =0.900, 95% CI: 0.839 to 0.960, P<0.001; Figure 4B).
Meta-analysis of miR-144-3p expression in HCC
Regarding the differential expression of miR-144-3p between HCC and nontumor liver tissues, six studies of microarrays in the GEO database (GSE41874, GSE40744, GSE21362, GSE22058, GSE12717, GSE57555), which involved 226 HCC tissues with 212 nontumor liver tissues, were included for this meta-analysis. Next, we combined the available data (TCGA, qRT-PCR, GEO) by a meta-analysis, which obtained the pooled SMD of miR-144-3p as −0.854 (95% CI: −1.224 to −0.484, P<0.001; Figure 5A) using a random-effects model, and the P-value of the heterogeneity test was <0.001 (I2=79%). Moreover, we performed a subgroup analysis which involved only six microarray studies, and a similar trend of SMD was also observed by a fixed-effects model (SMD =−0.608, 95% CI: −0.803 to −0.412, P<0.001; heterogeneity: I2=26%, P=0.239; Figure 5B). No obvious publication bias was noted (Begg’s test: P=0.707, Egger’s test: P=0.968; Figure 6A). Likewise, we did not find obvious publication bias among the included studies (Begg’s test: P=0.711, Egger’s test: P=0.654; Figure 6B).
Potential targets of miR-144-3p
Twelve databases were searched to predict the potential targets of miR-144-3p. A total of 4,297 genes reported in at least four databases were obtained from those databases, and 14 validated target genes with strong evidence were identified via the combination of the miRTarBase and TarBase databases. Next, the genes reported in at least four databases were combined with the validated target genes, and finally, 4,311 genes were selected to represent the targets of miR-144-3p. A flowchart of this prediction analysis is presented in Figure 7.
Overlap of the predicted target genes of miR-144-3p, the NLP analysis, the differential expression gene in HCC in TCGA data and its GO analysis
We extracted 1800 HCC-related genes in the NLP analysis and obtained 4,159 differential expression genes in HCC via the TCGA data. After combining the three aforementioned parts (the predicted targets of miR-144-3p, the NLP analysis for HCC-related genes and differential genes in HCC in TCGA data) by the intersection, 119 significant genes were identified to be responsible for the targets of miR-144-3p in the pathogenesis of HCC, and they were further utilized for the GO analysis. There were 264 significant terms involving biologic processes (P<0.05). Among them, we achieved response to organic substance, response to hormone stimulus, regulation of cell proliferation, response to endogenous stimulus and response to steroid hormone stimulus (Table 3). Moreover, there were 32 terms involving cellular components (P<0.05), some of which were associated with extracellular region, extracellular space, platelet alpha granule lumen, fibrinogen complex, cytoplasmic membrane-bounded vesicle lumen and so on (Table 3). Meanwhile, there were 26 significant terms enriched of molecular functions (P<0.05) including kinase binding, eukaryotic cell surface binding, cell surface binding, enzyme binding, sequence-specific DNA binding and so on (Table 3).
Furthermore, BINGO analysis was conducted for the visualization of the GO analyses based on the overlapping 119 genes. The associations among the GO terms are presented in Figure 8 (biologic processes), Figure 9 (cellular components) and Figure 10 (molecular function). The protein–protein interaction analysis was performed to identify the hub genes among the 119 overlapping genes, and in the protein–protein interaction analysis, we observed several genes in close association with the other genes, such as MAP2K1, JUN, SRC and CCND2 (Figure 11).
Pathway analysis of the overlapping genes between the predicted genes, the validated target genes of miR-144-3p and the NLP in KEGG
The overlapping 119 genes of the predicted genes, the NLP analysis and the differential expression of HCC in TCGA data were also applied to pathway analysis in KEGG database. There were 13 significant signaling pathways (P<0.05, Table 4), including pathways in cancer (AR, RET, MAP2K1, PTGS2, FOXO1, KITLG, GLI2, MMP1, CCNE2, CCNE1, FOS, ITGA6, CDKN2B, JUN, SLC2A1, PDGFRA, AXIN2, FIGF, P=3.73E-08); p53 signaling pathway (CCNE2, CCNB1, CCNE1, CCND2, CHEK1, THBS1, P=8.31E-04); cell cycle (CCNE2, CCNB1, CCNE1, CDKN2B, CCND2, CHEK1, MCM6, P=2.32E-03); focal adhesion (MAP2K1, ITGA6, CCND2, JUN, PDGFRA, THBS1, FIGF, SRC, P=6.08E-03); toll-like receptor signaling pathway (FOS, MAP2K1, JUN, IL1B, TLR4, 2.45E-02) and so on (Table 4).
Pathway analysis of the overlapping genes of the predicted genes of miR-144-3p, the NLP and the differentially expressed gene in HCC in TCGA data in Panther database
In the Panther database, there were four signaling pathways (P<0.05), including several vital pathways related to carcinogenesis and cancer progression, such as cell cycle (CCNE2, CCNB1, CCNE1, CCND2, P=1.15E-02) and angiogenesis (FOS, EPHA7, SFRP1, JUN, TEK, PDGFRA, AXIN2, ANGPT2, SRC, P=3.28E-02; Table 5).
Discussion
HCC, as a common cancer, attracts more attention in clinical practice worldwide because of the growing incidence and unsatisfactory survival despite all of the achievements in diagnosis and treatment.1,48 MiR-144-3p, a member of the miRNA family, can bind to its target mRNA in order to inhibit or promote oncogenesis and progression.49–51 Hence, it is pivotal to investigate and clarify the role of miR-144-3p in HCC in order to identify novel potential targets for the diagnosis, prediction of prognosis and treatment of HCC.
The expression of miR-144-3p has been explored in various cancers. Lower levels of miR-144-3p were found in breast cancer, renal cell carcinoma and hematologic malignancies.49,52,53 In esophageal carcinoma, Gao et al54 and Shao et al55 observed that miR-144-3p was underexpressed. However, Sharma et al reported an opposite pattern of miR-144-3p expression in esophageal carcinoma.56 The inconsistent findings in esophageal carcinoma may have resulted from the limited sample size. With regard to miR-144-3p expression in HCC, there have been only three individual studies which focused on miR-144-3p expression in HCC tissues with small sample size. Zhou et al,30 Cao et al31 and Yu et al29 unveiled that miR-144-3p was downregulated in 23 pairs, 33 pairs and 100 pairs of HCC tissues with corresponding normal control tissues, respectively. Likewise, we observed a decreased miR-144-3p level both in TCGA data and in qRT-PCR validation (both P<0.001). More importantly, a meta-analysis (a method of evidence-based medicine) was performed to accumulate the current individual evidence of the aberrant miR-144-3p in HCC, and we aimed to reach a more stable and convincing conclusion with a larger sample size. By combining the TCGA data, the qRT-PCR data and the GEO data, we confirmed the downregulation of miR-144-3p in 675 cases of HCC as compared to 357 normal liver tissues (SMD =−0.854, 95% CI: −1.224 to −0.484, P<0.001), and the result was more convincing than previous studies. After this, we also conducted the ROC curve to assess the diagnostic capability of miR-144-3p for HCC. Two significant AUCs of the ROC curves were reached in TCGA data (AUC =0.852, 95% CI: 0.810 to 0.894, P<0.001) and qRT-PCR validation (AUC =0.867, 95% CI: 0.817 to 0.916, P<0.001). Interestingly, after separating the cohort for qRT-PCR validation into two groups (AFP-positive HCC and AFP-negative HCC), the diagnostic value of miR-144-3p for HCC changed (AFP-positive HCC: AUC =0.827, 95% CI: 0.749 to 0.905, P<0.001; AFP-negative HCC: AUC =0.900, 95% CI: 0.839 to 0.960, P<0.001). Based on the results above, we demonstrated that miR-144-3p, to a certain extent, had diagnostic value for HCC, especially for AFP-negative HCC. To our knowledge, about 80% patients with HCC have elevated AFP in the serum. AFP is a clinically accepted biomarker for the diagnosis and recurrence of HCC. The AFP-negative HCC, without obvious clinical manifestations, is difficult to be diagnosed in the early stage. In our study, miR-144-3p showed considerable diagnostic value for AFP-negative HCC, which could provide a novel strategy to improve the early diagnosis of AFP-negative HCC patients. Further in vivo and in vitro investigations are required to clarify the relevant mechanisms of alterations in miR-144-3p expression in HCC.
Regarding the clinicopathologic significance of miR-144-3p in HCC, only Yu et al carried out the analysis unveiling that downregulated miR-144-3p was more remarkable in advanced TNM stage compared to early TNM stage.29 In this study, we observed miR-144-3p level was lower in metastasis (P=0.006), advanced TNM stage (P=0.001) and noncapsule or infiltration (P=0.047) groups than in nonmetastasis, early TNM stage and complete capsule groups, respectively. Notably, the aforementioned clinicopathologic features, which showed the differential expression of miR-144-3p, correlated well with the prognosis of HCC. Several published studies which investigated the regulatory mechanisms of miR-144-3p in HCC confirmed the relationships between miR-144-3p and unfavorable clinicopathologic features. Cao et al ascertained miR-144-3p could suppress the cell proliferation, migration and invasion in HCC by directly targeting E2F transcription factor 3 in vitro.31 From the perspective of the biologic process of miRNA with multiple targets, Yu et al clarified another target (SMAD family member 4, SMAD4) of miR-144-3p in HCC and showed that miR-144-3p could increase the chemosensitivity of 5-fluorouracil in HCC cell line.29 Similarly, Zhou et al elucidated that miR-144-3p could enhance the chemosensitivity through the regulation of Nrf2-dependent antioxidant pathway.30 These targets (E2F3, SMAD4 and Nrf2) of miR-144-3p validated in previous studies with strong evidence did not arise in this analysis. The reason may be that though the comprehensive bioinformatic investigation could provide relatively complete targets of miR-144-3p in HCC, the major limitation of bioinformatics was that the targets were identified with less evidence. Both false-positive and false-negative targets of miR-144-3p were inevitable. Thus, considering the complex mechanisms of miRNA and the pathogenesis of HCC, further in vitro and in vivo investigations are necessary to clarify the molecular mechanisms of miR-144-3p in HCC. Taken together, miR-144-3p may be a novel marker to predict the prognosis of HCC. Further survival analysis is essential to confirm the prognostic role of miR-144-3p.
To further elucidate the potential regulatory functions of miR-144-3p in carcinogenesis and the progression of HCC, we performed a bioinformatics analysis of the potential target genes of miR-144-3p in HCC. Three sections, 12 prediction databases and 2 validation databases for obtaining miR-144-3p targets, the NLP analysis searching for HCC-related genes and the differential expression of genes of HCC in TCGA database, were employed. The potential targets of miR-144-3p in the pathogenesis of HCC were identified by the overlapping among the three sections. In the GO analysis of the overlapping genes, some of the targets were associated with the regulation of cell proliferation, kinase binding, sequence-specific DNA binding and so on. These relevant functions of the overlapping genes may be indispensable for the pathogenesis in HCC. Further in vitro and in vivo experiments are also required to clarify the functions of these target genes of miR-144-3p in HCC. The protein-to-protein network of the overlapping genes revealed several hub genes which have been investigated in the carcinogenesis and progression, including MAP2K1, JUN, SRC, CCND2 and so on.57–60 Further in vivo and in vitro experiments are necessary to affirm the relationships between miR-144-3p and the hub genes in the pathogenesis of HCC.
In the pathway analysis of the overlapping genes, 13 noteworthy signaling pathways in the KEGG database and 4 significantly signaling pathways in the Panther database were enriched, including the p53 signaling pathway, the toll-like receptor signaling pathway and angiogenesis. Among them, several dysfunctional signaling pathways have been observed to contribute to oncogenesis and the progression of HCC.61–63 Further in vitro and in vivo exploration of miR-144-3p’s regulatory mechanism in these signaling pathways is imperative to confirm the role of miR-144-3p in HCC.
In conclusion, miR-144-3p may act as a tumor-suppressing miRNA in the carcinogenesis and progression of HCC via the regulation of cell proliferation, kinase binding and sequence-specific DNA binding, including several important pathways in the carcinogenesis and progression in the HCC: p53 signaling pathway, the toll-like receptor signaling pathway and the angiogenesis pathway. Further studies are necessary to verify the relationships between miR-144-3p and the key hub genes in the pathogenesis of HCC. Thus, interactions with miR-144-3p may provide an original insight for HCC treatment.
Acknowledgments
The study was supported partly by the Fund of National Natural Science Foundation of China (NSFC81560386), Guangxi Natural Science Fund for Innovation Research Team (2016GXNSFGA380006) and the Fund of Guangxi Zhuang Autonomous Region University Student Innovative Plan (No 201610598040). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. The authors thank TCGA and GEO for the public data.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Lin J, Wu L, Bai X, et al. Combination treatment including targeted therapy for advanced hepatocellular carcinoma. Oncotarget. 2016;7(43):71036–71051. | ||
Margini C, Dufour JF. The story of HCC in NAFLD: from epidemiology, across pathogenesis, to prevention and treatment. Liver Int. 2016;36(3):317–324. | ||
Zhao J, Zhang H, Wei L, Xie S, Suo Z. Comparing the long-term efficacy of standard and combined minimally invasive procedures for unresectable HCC: a mixed treatment comparison. Oncotarget. 2017;8(9):15101–15113. | ||
Balogh J, Victor D 3rd, Asham EH, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma. 2016;3:41–53. | ||
Qi X, Wang D, Su C, Li H, Guo X. Hepatic resection vs transarterial chemoembolization for the initial treatment of hepatocellular carcinoma: a systematic review and meta-analysis. Oncotarget. 2015;6(21):18715–18733. | ||
Zhang YF, Ho M. Humanization of high-affinity antibodies targeting glypican-3 in hepatocellular carcinoma. Sci Rep. 2016;6:33878. | ||
Colagrande S, Inghilesi AL, Aburas S, Taliani GG, Nardi C, Marra F. Challenges of advanced hepatocellular carcinoma. World J Gastroenterol. 2016;22(34):7645–7659. | ||
Nakamoto Y. Promising new strategies for hepatocellular carcinoma. Hepatol Res. 2017;47(4):251–265. | ||
Chauhan R, Lahiri N. Tissue- and serum-associated biomarkers of hepatocellular carcinoma. Biomark Cancer. 2016;8(Suppl 1):37–55. | ||
Zheng JF, Lu J, Wang XZ, Guo WH, Zhang JX. Comparative metabolomic profiling of hepatocellular carcinoma cells treated with sorafenib monotherapy vs. sorafenib-everolimus combination therapy. Med Sci Monit. 2015;21:1781–1791. | ||
Jianyong L, Jinjing Z, Jingcheng H, et al. Hepatocellular carcinoma cases with high levels of c-Raf-1 expression may benefit from postoperative adjuvant sorafenib after hepatic resection even with high risk of recurrence. Oncotarget. 2016;7(27):42598–42607. | ||
Tang S, Tan G, Jiang X, et al. An artificial lncRNA targeting multiple miRNAs overcomes sorafenib resistance in hepatocellular carcinoma cells. Oncotarget. 2016;7(45):73257–73269. | ||
Mizuguchi Y, Takizawa T, Yoshida H, Uchida E. Dysregulated miRNA in progression of hepatocellular carcinoma: a systematic review. Hepatol Res. 2016;46(5):391–406. | ||
Liu LN, Li DD, Xu HX, Zheng SG, Zhang XP. Role of microRNAs in hepatocellular carcinoma. Front Biosci (Landmark Ed). 2015;20:1056–1067. | ||
Liu L, Ye JX, Qin YZ, Chen QH, Ge LY. Evaluation of miR-29c, miR-124, miR-135a and miR-148a in predicting lymph node metastasis and tumor stage of gastric cancer. Int J Clin Exp Med. 2015;8(12):22227–22236. | ||
Jin K, Li T, Sanchez-Duffhues G, Zhou F, Zhang L. Involvement of inflammation and its related microRNAs in hepatocellular carcinoma. Oncotarget. 2017;8(13):22145–22165. | ||
Lee YS, Kim H, Kim HW, et al. High expression of MicroRNA-196a indicates poor prognosis in resected pancreatic neuroendocrine tumor. Medicine (Baltimore). 2015;94(50):e2224. | ||
Zhang X, Tang W, Chen G, et al. An encapsulation of gene signatures for hepatocellular carcinoma, MicroRNA-132 predicted target genes and the corresponding overlaps. PLoS One. 2016;11(7):e0159498. | ||
Zhang X, Tang W, Li R, et al. Downregulation of microRNA-132 indicates progression in hepatocellular carcinoma. Exp Ther Med. 2016;12(4):2095–2101. | ||
Wei Y, He R, Wu Y, et al. Comprehensive investigation of aberrant microRNA profiling in bladder cancer tissues. Tumour Biol. 2016;37(9):12555–12569. | ||
He R, Yang L, Lin X, et al. MiR-30a-5p suppresses cell growth and enhances apoptosis of hepatocellular carcinoma cells via targeting AEG-1. Int J Clin Exp Pathol. 2015;8(12):15632–15641. | ||
Brunetti O, Russo A, Scarpa A, et al. MicroRNA in pancreatic adenocarcinoma: predictive/prognostic biomarkers or therapeutic targets? Oncotarget. 2015;6(27):23323–23341. | ||
Li C, Chen X, Huang J, Sun Q, Wang L. Clinical impact of circulating miR-26a, miR-191, and miR-208b in plasma of patients with acute myocardial infarction. Eur J Med Res. 2015;20:58. | ||
Du DS, Yang XZ, Wang Q, et al. Effects of CDC42 on the proliferation and invasion of gastric cancer cells. Mol Med Rep. 2016;13(1):550–554. | ||
Giulietti M, Occhipinti G, Principato G, Piva F. Identification of candidate miRNA biomarkers for pancreatic ductal adenocarcinoma by weighted gene co-expression network analysis. Cell Oncol. 2017;40(2):181–192. | ||
Hua L, Zhou P, Li L, Liu H, Yang Z. Prioritizing breast cancer subtype related miRNAs using miRNA-mRNA dysregulated relationships extracted from their dual expression profiling. J Theor Biol. 2013;331:1–11. | ||
Yepes S, Lopez R, Andrade RE, Rodriguez-Urrego PA, Lopez-Kleine L, Torres MM. Co-expressed miRNAs in gastric adenocarcinoma. Genomics. 2016;108(2):93–101. | ||
Yu M, Lin Y, Zhou Y, et al. MiR-144 suppresses cell proliferation, migration, and invasion in hepatocellular carcinoma by targeting SMAD4. Onco Targets Ther. 2016;9:4705–4714. | ||
Zhou S, Ye W, Zhang Y, et al. miR-144 reverses chemoresistance of hepatocellular carcinoma cell lines by targeting Nrf2-dependent antioxidant pathway. Am J Transl Res. 2016;8(7):2992–3002. | ||
Cao T, Li H, Hu Y, Ma D, Cai X. miR-144 suppresses the proliferation and metastasis of hepatocellular carcinoma by targeting E2F3. Tumour Biol. 2014;35(11):10759–10764. | ||
Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS One. 2013;8(4):e61054. | ||
Rong M, He R, Dang Y, Chen G. Expression and clinicopathological significance of miR-146a in hepatocellular carcinoma tissues. Ups J Med Sci. 2014;119(1):19–24. | ||
Chen G, Umelo IA, Lv S, et al. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8(3):e60317. | ||
Huang WT, Wang HL, Yang H, et al. Lower expressed miR-198 and its potential targets in hepatocellular carcinoma: a clinicopathological and in silico study. Onco Targets Ther. 2016;9:5163–5180. | ||
Huang WT, Chen ZX, He RQ, et al. Clinicopathological role of miR-30a-5p in hepatocellular carcinoma tissues and prediction of its function with bioinformatics analysis. Onco Targets Ther. 2016;9:5061–5071. | ||
Liu Y, Ren F, Luo Y, Rong M, Chen G, Dang Y. Down-regulation of MiR-193a-3p dictates deterioration of HCC: a clinical real-time qRT-PCR study. Med Sci Monit. 2015;21:2352–2360. | ||
Liu Y, Ren F, Rong M, Luo Y, Dang Y, Chen G. Association between underexpression of microrna-203 and clinicopathological significance in hepatocellular carcinoma tissues. Cancer Cell Int. 2015;15:62. | ||
Gan TQ, Tang RX, He RQ, Dang YW, Xie Y, Chen G. Upregulated MiR-1269 in hepatocellular carcinoma and its clinical significance. Int J Clin Exp Med. 2015;8(1):714–721. | ||
Rong M, Chen G, Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. | ||
Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ. 2007;335(7626):914–916. | ||
Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. | ||
Vlachos IS, Paraskevopoulou MD, Karagkouni D, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43(Database issue):D153–D159. | ||
Chou CH, Chang NW, Shrestha S, et al. miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016;44(D1):D239–D247. | ||
Fan Y, Siklenka K, Arora SK, Ribeiro P, Kimmins S, Xia J. miRNet – dissecting miRNA-target interactions and functional associations through network-based visual analysis. Nucleic Acids Res. 2016;44(W1):W135–W141. | ||
Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. | ||
Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. | ||
Chacko S, Samanta S. “Hepatocellular carcinoma: a life-threatening disease”. Biomed Pharmacother. 2016;84:1679–1688. | ||
Pan Y, Zhang J, Fu H, Shen L. miR-144 functions as a tumor suppressor in breast cancer through inhibiting ZEB1/2-mediated epithelial mesenchymal transition process. Onco Targets Ther. 2016;9:6247–6255. | ||
Jin J, Wang Y, Xu Y, et al. MicroRNA-144 regulates cancer cell proliferation and cell-cycle transition in acute lymphoblastic leukemia through the interaction of FMN2. J Gene Med. 2016. | ||
Liu M, Gao J, Huang Q, Jin Y, Wei Z. Downregulating microRNA-144 mediates a metabolic shift in lung cancer cells by regulating GLUT1 expression. Oncol Lett. 2016;11(6):3772–3776. | ||
Liu F, Chen N, Xiao R, Wang W, Pan Z. miR-144–3p serves as a tumor suppressor for renal cell carcinoma and inhibits its invasion and metastasis by targeting MAP3K8. Biochem Biophys Res Commun. 2016;480(1):87–93. | ||
Cai SD, Chen JS, Xi ZW, Zhang LJ, Niu ML, Gao ZY. MicroRNA144 inhibits migration and proliferation in rectal cancer by downregulating ROCK1. Mol Med Reps. 2015;12(5):7396–7402. | ||
Gao Z, Liu R, Liao J, et al. Possible tumor suppressive role of the miR-144/451 cluster in esophageal carcinoma as determined by principal component regression analysis. Mole Med Rep. 2016;14(4):3805–3813. | ||
Shao Y, Li P, Zhu ST, et al. MiR-26a and miR-144 inhibit proliferation and metastasis of esophageal squamous cell cancer by inhibiting cyclooxygenase-2. Oncotarget. 2016;7(12):15173–15186. | ||
Sharma P, Saraya A, Sharma R. Potential diagnostic implications of miR-144 overexpression in human oesophageal cancer. Indian J Medi Res. 2016;143(Suppl):S91–S103. | ||
Janku F, Kaseb AO, Tsimberidou AM, Wolff RA, Kurzrock R. Identification of novel therapeutic targets in the PI3K/AKT/mTOR pathway in hepatocellular carcinoma using targeted next generation sequencing. Oncotarget. 2014;5(10):3012–3022. | ||
Yang Z, Zhang Y, Wang L. A feedback inhibition between miRNA-127 and TGFbeta/c-Jun cascade in HCC cell migration via MMP13. PLoS One. 2013;8(6):e65256. | ||
Tsutsui M, Iizuka N, Moribe T, et al. Methylated cyclin D2 gene circulating in the blood as a prognosis predictor of hepatocellular carcinoma. Clin Chim Acta. 2010;411(7–8):516–520. | ||
Zhao R, Wu Y, Wang T, et al. Elevated Src expression associated with hepatocellular carcinoma metastasis in northern Chinese patients. Oncol Lett. 2015;10(5):3026–3034. | ||
Huang Q, Li J, Xing J, et al. CD147 promotes reprogramming of glucose metabolism and cell proliferation in HCC cells by inhibiting the p53-dependent signaling pathway. J Hepatol. 2014;61(4):859–866. | ||
Dong YQ, Lu CW, Zhang L, Yang J, Hameed W, Chen W. Toll-like receptor 4 signaling promotes invasion of hepatocellular carcinoma cells through MKK4/JNK pathway. Mol Immunol. 2015;68(2 Pt C):671–683. | ||
Yang X, Zhang XF, Lu X, et al. MicroRNA-26a suppresses angiogenesis in human hepatocellular carcinoma by targeting hepatocyte growth factor-cMet pathway. Hepatology. 2014;59(5):1874–1885. | ||
Guo Y, Ying L, Tian Y, et al. miR-144 downregulation increases bladder cancer cell proliferation by targeting EZH2 and regulating Wnt signaling. FEBS J. 2013;280(18):4531–4538. | ||
Gu H, Liu M, Ding C, et al. Hypoxia-responsive miR-124 and miR-144 reduce hypoxia-induced autophagy and enhance radiosensitivity of prostate cancer cells via suppressing PIM1. Cancer Med. 2016;5(6):1174–1182. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.