Back to Journals » Infection and Drug Resistance » Volume 15
A Complete Genome of Nocardia terpenica NC_YFY_NT001 and Pan-Genomic Analysis Based on Different Sources of Nocardia spp. Isolates Reveal Possibly Host-Related Virulence Factors
Authors Cai Q, Huang Y , Zhou L, Hu N, Liu Y, Guo F , Liu Q, Huang X, Zhang Y, Zeng L
Received 16 August 2022
Accepted for publication 29 November 2022
Published 13 December 2022 Volume 2022:15 Pages 7259—7270
DOI https://doi.org/10.2147/IDR.S384673
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Qinming Cai,1,2,* Yongcheng Huang,1,* Li Zhou,1 Niya Hu,1 Yanling Liu,1 Fujia Guo,1 Qiong Liu,3 Xiaotian Huang,1,3 Yunyi Zhang,4 Lingbing Zeng1
1The First Affiliated Hospital of Nanchang University, School of Public Health, Nanchang University, Nanchang, People’s Republic of China; 2The Second School of Clinical Medicine, Southern Medical University, Guangzhou, People’s Republic of China; 3Department of Medical Microbiology, School of Medicine, Nanchang University, Nanchang, People’s Republic of China; 4Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention, Hangzhou, 310051, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yunyi Zhang; Lingbing Zeng, Email [email protected]; [email protected]
Objective: We aimed to identify the possible virulence genes associated with Nocardia NC_YFY_NT001 isolated by ourselves and other Nocardia spp.
Methods: The genome of Nocardia terpenica NC_YFY_NT001 was completed by using PacBio and Illumina platforms. A pan-genomic analysis was applied to selected complete Nocardia genomes.
Results: Nocardia terpenica NC_YFY_NT001 can cause healthy mice death by tail intravenous injection. The genome of NT001 has one circular chromosome 8,850,000 bp and one circular plasmid 70,000 bp with ~68% GC content. The chromosome and plasmid encode 7914 and 80 proteins, respectively. Furthermore, a pan-genomic analysis showed a total of 45,825 gene clusters, then 304 core, 21,045 shell and 24,476 cloud gene clusters were classified using specific parameters. In addition, we found that catalases were more abundant in human isolates. Furthermore, we also found no significant differences in the MCE proteins between different strains from different sources. The pan-genomic analysis also showed that 67 genes could only be found in humoral isolates. ReX3 and DUF853 domain protein were found in all eight human isolates. The composition of unique genes in humoral isolate genomes indicated that the transcriptional regulators may be important when Nocardia invades the host, which allows them to survive in the new ecological system.
Conclusion: In this study, we confirmed that NT001 could cause infected animal death, and identified many possible virulence factors for our future studies. This study also provides new insight for our further study on Nocardia virulence mechanisms.
Keywords: Nocardia, virulence, pan-genome, infection
Introduction
Actinobacteria belong to a phylum of Gram-positive bacteria that provide important contributions to soil systems.1 Many actinobacteria are branched and always grow extensive mycelia. According to their special characteristics, most of actinobacteria are important in bioproducts synthesis, including immunosuppressive compounds, cytostatic and antibiotic. Both Mycobacteriaceae and Nocardiaceae belong to Corynebacterineae, and can produce mycolic acids, which can help pathogens resistant to the attack of hosts. Nocardia belongs to Nocardiaceae family, which can cause immunocompromised patient diseases.2 The genus of Nocardia is Gram-positive and catalase positive and comprises aerobic bacteria that belong to actinobacteria.3–5 Corynebacterium, Mycobacterium and Nocardia form the well-known CMN actinobacterial group. Similar to other actinobacteria, mycolic acids in the cell wall represent one of the important structures of Nocardia, making them act as partially acid-fast bacteria.6 Nocardia is not a commensal bacterium, and was first described by Edmond Nocard in 1888.7 With the rising number of immunocompromised patients, nocardiosis incidence has recently increased, but the pathogenic mechanisms of nocardiosis are still unclear.8–10
Nocardial species can cause pulmonary, cutaneous and system infections in humans, one of the most frequent being pulmonary nocardiosis.11 N. farcinica, N. cyriacigeorgica, N. brasiliensis, and N. nova are the most frequent isolates.12 Another common infection is CNS nocardiosis in immunocompromised patients. N. abscessus, N. araoensis, N. asiatica, N. asteroids, N. brasiliensis, N. caviae, N. cerradoensis, N. crassostreae, N. cyriacigeorgica, N. farcinica, N. mexicana, N. nova, N. otitidiscaviarum, N. paucivorans, N. pseudobrasiliensis and N. veterana have all been isolated in CNS-infected patients,13–24 although in most of the cases, the patients were dead. Interestingly, we isolated a new nocardial strain from a bacteriemia and CNS-infected patient, N. terpenica NC_YFY_NT001.25 N. terpenica was first identified in 2007 in a patient suffering from pulmonary nocardiosis.26 Therefore, it was the first example of N. terpenica causing CNS infection, and the patient eventually recovered.25 The experimentally verified virulence factors including genes encoding catalase, cell-wall lipids, superoxide dismutase, hydrolases, phospholipase C, hemolysin (toxic proteins), lipases and proteases have been identified in Nocardia spp. (Nocardia brasiliensis, Nocardia farcinica, Nocardia cyriacigeorgica).2,27 Catalase and superoxide dismutase (SOD) block the function of phagocytes to degrade the bacteria, while phospholipase C has the function of destroying tissue.28 In order to better understand the pathogenic mechanisms of N. terpenica, we determined the entirety of the genome of NT001, and found some potential virulence factors, such as catalase, MCE proteins and lipases, which were are consistent with the results reported in previous studies.
In the past few decades, the genomes of several nocardial species have been sequenced, and since we updated our genome, 12 nocardial genomes from different environments, including human, have been completed. Unfortunately, few studies have focused on understanding genomic differences from various sources.29,30 Therefore, we finished a pan-genomic analysis of nocardial species by using the completed genomes. Our aims were to characterize the potential factors related to human infection and provide enough additional information for nocardiosis diagnosis in future.
Materials and Methods
Strain Isolation and Culture
N. terpenica NC_YFY_NT001 was isolated from a bacteremia-infection patient, and the case was published in our previous study. The isolation process could be concised as follows: the patient was admitted to the hospital as fever and headache. Then, Nocardia was isolated from blood and cerebrospinal fluid, and identified by 16s rRNA sequencing. In this study, we cultured the Nocardia using brain-heart infusion broth (BHI-P), at 35°C for 72h.
Animal Experimentation
Female BALB/c mice weighing around 18–20 g (approximately 8 weeks old) were maintained by the Laboratory Animal Center, Nanchang University in accordance with protocols approved by the university (approval No. NCUSYDWFL-201035). This study was performed in line with the principles of the Declaration of Helsinki or comparable ethical standards.
N. terpenica NC_YFY_NT001 strain was grown in brain–heart infusion broth (BHI-P) to mid-log phase at 37 °C with intense rotational agitation (220 rpm). To avoid bacterial clumps, the culture broth was settled on the top for ~ 30 minutes, and the supernatant was adjusted at approximately 5 × 108 CFU/mL. Then, according to detailed description in the previous study, 0.1mL this suspension was injected intravenously into each mouse through the lateral caudal vein (IV), in which each mouse received about 5 × 107CFU. The animal experiment was repeated three times, and five mice per group. Lungs, hearts, livers and kidneys were removed from the mice after one-week infection, then fixed with 4% paraformaldehyde. Afterwards, tissues were dehydrated, imbedded in paraffin wax, and cut into 2 μm-thick sections. Routine histological staining methods were used.
Genome Sequencing and Assembly
N. terpenica NC_YFY_NT001 was sequenced by using the PacBio and HiSeq platforms. For the Illumina HiSeq, genomic DNA (1 ug) was sheared using Covaris S2 (Covaris, USA), then agarose gel electrophoresis and a 300 bp fragment was recovered and purified by gelatinization. A TruSeqTM DNA Sample Prep Kit—Set A (llumine, USA) and TruSeq PE Cluster Kit (Illumina, USA) were used for the 300 bp-insert PE library construction, and were finally sequenced on an Illumina HiSeq Sequencer. For the PacBio RS II platform, genomic DNA (5 ug) was sheared with a g-tube, 8–12 K DNA fragment purification with AMPure beads (Beckman Coulter, USA), and a 10 K template library was generated by using a DNA Template Prep Kit 2.0 (Pacific Biosciences, USA); then, the genomes were sequenced on the PacBio RSII platform (PacBio).
We filtered the PacBio polymerase reads and obtained the subread; then, HGAP software was subsequently applied for the assembly results. The Illumina data were mapped to the genome using Bowtie2 software.31,32
Genome Annotation and Analysis
The genome sequence was automatically annotated using the NCBI Prokaryotic Genome Annotation Pipeline. tRNA genes were recognized by tRNAscan, and rRNA genes were predicted via RNAmmer. Clusters of Orthologous Groups (COG) classification was used for the CDD database, and the construction of metabolic pathways via the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The genome sequence of N. terpenica YFY_NC_NT_001 is publicly available at https://www.ncbi.nlm.nih.gov/genome/14487?genome_assembly_id=343105. The sequences and annotations data have been deposited at the NCBI database and given the accession number NZ_CP023778.
Prophage Analysis
PHASTER tool was used for predicting Nocardia prophage with the default dataset.33
Pan-Genomic Analysis
Sequence data for comparative analyses were obtained from the NCBI database. IslandViewer was used for genomic island prediction. Comparative genomics analysis was carried out by using the Roary pipeline (identity=85), and Scoary was used for analyzing the specific genes in human or environment.34 Virulence factors were predicted by VFDB (identity > 50%, coverage ≥ 20%).35
Results and Discussion
Virulence of N. Terpenica NC_YFY_NT001
N. terpenica NC_YFY_NT001 was isolated from a diabetes patient, who was under an immunocompromised condition.25 In order to verify the virulence of N. terpenica NC_YFY_NT001, the strain was tested in BALB/c mice by tail vein injection using 5×107 bacteria; more than 60% mice were dead at day 3. Then, the degree of pathological damage was evaluated by H&E staining. The pathological results were confirmed by two different doctors. It showed an obvious destruction of alveoli in our experimental groups, and we could identify scattered lymphocytic infiltration. In the liver, unclassical proliferation of liver cells was found; in the spleen, proliferation of lymphocytes was found. There were no significant differences in the kidneys (Figure 1). Previous study reported that most individuals with Nocardia colonization had a lung disease, which may be related to the fact that lower respiratory tract epithelial damage can facilitate the presence of Nocardia.36,37 The outcome of pulmonary nocardiosis is poor, with a 1-year survival rate of 55.4%.38 Indeed, we considered that NT001 could cause death through destruction of the lungs in the mice.
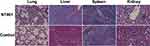 |
Figure 1 Hematoxylin and eosin-stained histologic sections of rat lung, liver, spleen, and kidney. |
General Characteristics of the N. Terpenica NC_YFY_NT001 Genome
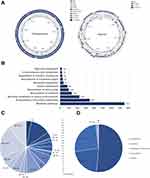
Due to a lack of studies on N. terpenica, the virulence factors and pathogenic process of the bacteria were unclear. Therefore, we finished the genome by combining it with the PacBio and HiSeq platforms. The genome of N. terpenica NC_NT001 contains one chromosome and one plasmid after assembly. Table 1 summarizes the genomic characteristics of NT001, and Figure 2A presents a physical map of the NT001 genome. In Table 1, we summarize the size of the NT001 chromosome, which is 8,850,000 bp, and the plasmid, which is 70,000 bp, with a GC% of ~68%. The chromosome genome encodes 56 tRNA, 9 rRNA, 3 other RNA and 7,914 genes. Pseudogenes were also predicted to be 228 in the chromosome. The plasmid has 80 genes and 2 pseudo genes. Most CDSs (about ~79%) have a function annotation. The other CDSs could be annotated as “hypothetical proteins”.
 |
Table 1 Genomic Information of N. terpenica NC_NT001 |
 |
Table 2 Genomic Information of Nocardial Strains for Pangenomic Analysis |
The functional gene categories comparison of different Nocardia was analyzed by BLAST P. With COG and KEGG pathway analysis, proteins of NT001 could be divided into 22 COG categories and 23 different KEGG pathways (Figure 2B and C). The gene abundance of “Secondary metabolites biosynthesis, transport and catabolism” (Q, 7.23%), “General function prediction only” (R, 13.27%) and “Function unknown” (S, 7.19%) were higher than other Nocardia. Interestingly, the abundance of genes encoded “Coenzyme metabolism” (H, 4.9%), “Lipid metabolism” (I, 6.02%), “Translation, ribosomal structure and biogenesis” (J, 3.83%) and “Inorganic ion transport and metabolism” (P, 3.67%) were much lower than other Nocardia species. This indicated that the pathogenic process of NT001 should be different from other Nocardia, and it is important to understand the function of those “Function unknown” proteins.
Furthermore, as extracellular proteins are the important virulence factors in bacteria, we predicted the subcellular localization of NT001 proteins using psortb 2.0 (Figure 2D). The results indicated that NT001 had 127 extracellular proteins. Many putative virulence factors were shown in the extracellular proteins, such as esterase, invasin and mycoltransferase.
Genomic Islands (GIs) Analysis
Genomic islands are important gene transfer elements in bacterial genomes and are abundant in many organisms. In our study, we could observe several horizontal gene transfer events in the NT001 genome. About 15 GIs were predicted to be present in the genome of NT001. The 15 GIs comprised 417 genes, and about 267 genes were annotated as hypothetical proteins. Supplementary S1 summarizes the categories of the NT001 island genes, including sources for genes encoding for defense, replication, recombination and repair, lipid/coenzyme/carbohydrate/nucleotide transport and metabolism, secondary metabolite biosynthesis, transport and catabolism, inorganic ion transport and metabolism, posttranslational modification, protein turnover, chaperones, cell cycle control, cell division, chromosome partitioning, and energy production and conversion. Many transposases were predicted to be components of GIs, which are related to horizontal gene transfer in the bacteria, and were scattered throughout the genome. Many phage-related genes were also found in the genome of NT001. Furthermore, ESX secretion-associated protein EspG, alpha/beta hydrolase, esterase and many transcriptional regulators are associated with GIs. These genes might be related to the pathogenic process of nocardiosis. It can be concluded that NT001 could receive some virulence factors through GIs, and could adapt to an in vivo environment. Prophage is another important gene transfer element in the bacteria, and is a bacteriophage genome that is important for regulating the microbial population density. PHAST were used for predicting the presence of prophage in the genome of NT001. The result indicated that NT001 harbored one questionable prophage. The size of this prophage is 26.9 kb, and it accounted for 0.3% of the genome (Supplementary S1). Overall, gene transfer is one of the important ways to swap genetic information between different microbial organisms, and the occurred event would be better for pathogens adapting to the host environment.
Pan-Genomic Construction of Nocardia spp.
At the time we submitted the genome of N. terpenica, 11 other Nocardial genomes were finished isolated from different sources (Table 2). Eight Nocardia strains were isolated from humans, and the rest were isolated from animals or soil. Most of the isolates had an ~8 Mbp chromosome, except Nocardia farcinica NCTC11134, whose chromosome was only 3.65 Mbp. Previous studies indicated that the size of the genome would have no clear correlation with virulence of Nocardia, which is consistent with our results.39 In addition, the genomes of Nocardia varied greatly, despite belonging to the same species. Like N. farcinica IFM10152, N. farcinica NCTC11134 and N. farcinica W6977, the composition of the genomes was different. IFM10152 had four plasmids, but the other two had only two plasmids. Furthermore, the chromosome size of these three strains was different as well. IFM10152 had a 3.65 Mbp chromosome, but the other two had a chromosome with ~6 Mbp. In order to determine the key factors related to infected humans, we carried out a pan-genomic analysis among these Nocardia species because of their different isolation sources.
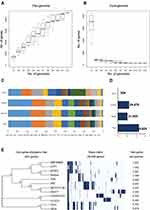
To investigate the differences among strains of Nocardia, a pan-genomic analysis of genomes of 12 different strains was performed. Prior to the analysis, the genomes were re-annotated by Prokka. The whole genome of Nocardia was used to construct the Nocardia database. Analysis was performed using the Roary pipeline. According to the gene accumulation curve, Nocardia showed an open genome structure, which increased in size with the number of genomes added, containing 45,825 gene clusters (Figure 3A and B, Supplementary S2).
Only 304 (0.66%) gene clusters were classified as core genes, and 21,045 gene clusters were identified as shell genes (45.93%). The remaining 24,476 (53.41%) gene clusters were classified as cloud genes (Figure 3C and D).
Functional Genomic Analysis
In order to obtain more information about the functional characteristics of Nocardia pan-genome, we use COG mapping to the eggNOG database to characterize the functions of core, shell and cloud genes.
To gain more information of the functional features of the Nocardia pan-genome, we characterized the functions of the core, shell and cloud genes by COG mapping to the eggNOG database. The highest proportion (24.11%) of the pan-genome was characterized as “S: function unknown”, which indicated that these genes had low similarity to the proteins in the database. Similar to pan-genome, shell (25.03%) and cloud (23.4%) genes had a high proportion of “function unknown (S)”. Genes assigned to “K: transcription” (1402 genes), “E: Amino acid transport and metabolism” (907 genes), “L: Lipid transport and metabolism” (787 genes), “C: Energy production and conversion” (676 genes) and “Q: Secondary metabolites biosynthesis, transport and catabolism” (561 genes) were prominently represented in the shell component of this pan-genome. Most genes were classified as “S: Function unknown” (1905 in cloud genes and 2,445 in shell genes). In addition, cloud genes were mostly enriched in “K: Transcription” (1265 genes), “I: Lipid transport and metabolism” (754 genes), “L: Replication, recombination and repair” (639 genes), “E: Amino acid transport and metabolism” (619 genes), “C: Energy production and conversion” (601 genes). Genes assigned to “K: transcription” (1402 genes), “E: Amino acid transport and metabolism” (907 genes), “I: Lipid transport and metabolism” (787 genes), “C: Energy production and conversion” (676 genes) and “Q: Secondary metabolites biosynthesis, transport and catabolism” (561 genes) were dominant in the shell component of pan-genomic analysis. The core genome was different from the cloud and shell genomes. The core genome was enriched in “J: Translation, ribosomal structure and biogenesis” (48 genes), “S: Function unknown” (40 genes), “E: Amino acid transport and metabolism” (37 genes) and “K: transcription” (33 genes).
Cloud gene and shell gene belong to non-core genes, which are genes present in one or more strains. We identified that shell (25.03%) and cloud (23.4%) genes had a high proportion of “function unknown (S)”, indicating that these genes had low similarity to the proteins in the database. These data suggested that the biological behavior and pathogenesis of N. terpenica YFY NC NT001 might be different from that of Nocardia strains found in the past. K (transcription) and E (amino acid transport and metabolism) play an important role in the growth and colonization of Nocardia, so they account for a relatively large proportion in all COG classification, whether in cloud gene, shell gene or core gene.40 The core genome is different from the variable genome (cloud and shell genomes), representing genes that exist in all strains. In our study, core genome was enriched in “J: Translation, ribosomal structure and biogenesis” (48 genes), “S: Function unknown” (40 genes), “E: Amino acid transport and metabolism” (37 genes) and “K: transcription” (33 genes). Most of the COG categories are known, indicating that N. terpenica YFY NC NT001 is a strain of Nocardia.
Furthermore, a core gene alignment tree was constructed by using the Mega N-J method. Interestingly, NT_001 and animal isolated Nocardia could be assigned as the same ancestors. The respiratory isolates seemed to be assigned as the same ancestors as soil isolate (Figure 3E). It indicated that the different sources of Nocardia might have different targets.
Potential Virulence Factors According to Pan-Genome
Until now, several studies have reported Nocardia virulence factors, including catalases, toxin-antitoxin (TA) systems, hemolysins, invasins and MCE proteins.41,42 Catalase is an important virulence factor that could inhibit the function of H2O2, especially during the process of escaping from phagocyte.43 As shown in Figure 4A, the genes of catalases in humoral isolates were more than non-humoral isolates (Table 3). The results indicated that catalase might be the vital factor that influences Nocardial species-infected humans. Catalase, Superoxide dismutase, Phospholipase C, Putative invasion, Proteases, Clpprotease, PE-PPE, Mce family protein, Metalloprotease, Salicylate synthase, Polyketide synthase, and Mycolyltransferase were detected as common virulence genes in our study, which were consistent with those reported in the past.
 |
Figure 4 Comparisons of virulence-associated genes in Nocardia spp. (A) Putative virulence factors in different Nocardia spp.; (B) Toxin–antitoxin-related genes in different Nocardia spp. |
 |
Table 3 Virulence Factors in Different Nocardial Strains |
Toxin-antitoxin (TA) systems are small genetic elements that focus on balancing the prokaryotes’ survival. However, there were no significant differences between the various sources of Nocardia spp. (Figure 4B). Hemolysins are another important virulence factor that could influence the bacterial pathogenic process. The NT001 genome encoded one hemolysin. Meanwhile, we also compared the hemolysins in different nocardial genomes and found that there was no hemolysin homolog in N. farcinica genomes. In this study, we also detected some other potential virulence factors that related to nocardiosis. Most Nocardia-related virulence factors were found in the genome of NT001, and the amounts of the genes are shown in Figure 4A.
In addition, we used Scoary to search for the genes only exist in human isolates. Finally, we found 67 genes could be found only in human isolates, and there were two proteins could be found in all eight human isolates: regX3 and hypothetical protein (group 1193, DUF853 protein). RegX3 belongs to a two components regulatory system, SenX3-RegX3, which controls the phosphate acquisition. And regX3 is an important virulence in the Mycobacteria.44–46 Therefore, this gene might be important during Nocardia spp. infect human beings. However, the function of DUF853 has no report until now. It is worth to figure out its function in our next study to elucidate whether it influence the virulence of Nocardia spp. or not.
Conclusions
We isolated a N. terpenica strain from a bacteremia-infection patient. In order to better understand the virulence factors and pathogenic mechanisms of the strain, we finished the genome of N. terpenica NC_YFY_NT001. The genome comprises a circular chromosome and a circular plasmid. The chromosome is 8,850,000 bp, and the plasmid is 70,000 bp, with a GC% of ~68%. Several genes involved the virulence of nocardiosis, such as catalase, MCE proteins and lipases. A pan-genomic analysis of Nocardia spp. also showed that some genes, regX3 and DUF853 domain protein were associated with the process of invading and adapting in humans, which warrants further investigation.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 32060040), Projects of Jiangxi Natural Science Foundation (20192ACBL21042, 20202BAB216045 and 20204BCJL23054).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Goodfellow M, Cross T. The Biology of the Actinomycetes. Academic Press; 1985:541.
2. Vera-Cabrera L, Ortiz-Lopez R, Elizondo-Gonzalez R, Ocampo-Candiani J, Gordon SV. Complete genome sequence analysis of Nocardia brasiliensis HUJEG-1 reveals a saprobic lifestyle and the genes needed for human pathogenesis. PLoS One. 2013;8(6):e65425. doi:10.1371/journal.pone.0065425
3. Wilson JW. Nocardiosis: updates and clinical overview. Mayo Clin Proc. 2012;87(4):403–407. doi:10.1016/j.mayocp.2011.11.016
4. Kurup PV, Randhawa HS, Sandhu RS. A survey of Nocardia asteroides, N. caviae and N. brasiliensis occuring in soil in India. Sabouraudia. 1968;6(3):260–266. doi:10.1080/00362176885190491
5. Han HJ, Kwak MJ, Ha SM, et al. Genomic characterization of Nocardia seriolae strains isolated from diseased fish. Microbiologyopen. 2019;8(3):e00656. doi:10.1002/mbo3.656
6. Nishiuchi Y, Baba T, Hotta HH, Yano I. Mycolic acid analysis in Nocardia species. The mycolic acid compositions of Nocardia asteroides, N. farcinica, and N. nova. J Microbiol Methods. 1999;37(2):111–122. doi:10.1016/s0167-7012(99)00055-x
7. Nocard E. Note sur la maladie des boeufs de la Guadeloupe connue sous le nom de farcin. Ann de l’Institut Pasteur. 1888;2:293–302.
8. Long PF. A retrospective study of Nocardia infections associated with the acquired immune deficiency syndrome (AIDS). Infection. 1994;22(5):362–364. doi:10.1007/BF01715551
9. Ali T, Chakraburtty A, Mahmood S, Bronze MS. Risk of nocardial infections with anti-tumor necrosis factor therapy. Am J Med Sci. 2013;346(2):166–168. doi:10.1097/MAJ.0b013e3182883708
10. Wadhwa T, Baveja U, Kumar N, Govil D, Sengupta S. Clinical manifestations of nocardiosis: study of risk factors and outcomes in a tertiary care hospital. J Lab Physicians. 2017;9(4):288–295. doi:10.4103/JLP.JLP_111_16
11. Brown-Elliott BA, Brown JM, Conville PS, Wallace RJ. Clinical and laboratory features of the Nocardia spp. based on current molecular taxonomy. Clin Microbiol Rev. 2006;19(2):259–282. doi:10.1128/CMR.19.2.259-282.2006
12. Minero MV, Marin M, Cercenado E, Rabadan PM, Bouza E, Munoz P. Nocardiosis at the turn of the century. Medicine. 2009;88(4):250–261. doi:10.1097/MD.0b013e3181afa1c8
13. Eisenblatter M, Disko U, Stoltenburg-Didinger G, et al. Isolation of Nocardia paucivorans from the cerebrospinal fluid of a patient with relapse of cerebral nocardiosis. J Clin Microbiol. 2002;40(9):3532–3534. doi:10.1128/jcm.40.9.3532-3534.2002
14. Jeong JH, Moon SM, Park PW, et al. Multiple brain abscesses caused by nocardia asiatica in a patient with systemic lupus erythematosus: the first case report and literature review. Ann Lab Med. 2017;37(5):459–461. doi:10.3343/alm.2017.37.5.459
15. Kennedy KJ, Chung KH, Bowden FJ, et al. A cluster of nocardial brain abscesses. Surg Neurol. 2007;68(1):43–9; discussion 49. doi:10.1016/j.surneu.2006.08.067
16. Bradsher RW, Monson TP, Steele RW. Brain abscess due to Nocardia caviae. Report of a fatal outcome associated with abnormal phagocyte function. Am J Clin Pathol. 1982;78(1):124–127. doi:10.1093/ajcp/78.1.124
17. Piau C, Kerjouan M, Le Mouel M, et al. First case of disseminated infection with Nocardia cerradoensis in a human. J Clin Microbiol. 2015;53(3):1034–1037. doi:10.1128/JCM.02979-14
18. Igbaseimokumo U, El Shafie S, Al Khal AL. First human infection of nocardia crassostreae in an immunocompetent patient. Chin Med J. 2016;129(1):114–115. doi:10.4103/0366-6999.172609
19. Barnaud G, Deschamps C, Manceron V, et al. Brain abscess caused by Nocardia cyriacigeorgica in a patient with human immunodeficiency virus infection. J Clin Microbiol. 2005;43(9):4895–4897. doi:10.1128/JCM.43.9.4895-4897.2005
20. Yamamoto F, Yamashita S, Kawano H, et al. Meningitis and ventriculitis due to Nocardia araoensis infection. Intern Med. 2017;56(7):853–859. doi:10.2169/internalmedicine.56.7332
21. Majeed A, Mushtaq A, Zangeneh T, et al. Intractable cerebral Nocardia mexicana in a GvHD patient successfully treated with linezolid. Bone Marrow Transplant. 2017;52(10):1476–1478. doi:10.1038/bmt.2017.167
22. Gezici AR, Daglioglu E, Ergungor F, Okay O, Polat O. Cerebral abscess caused by Nocardia nova. Neurol Neurochir Pol. 2008;42(2):153–156.
23. Brown BA, Lopes JO, Wilson RW, et al. Disseminated Nocardia pseudobrasiliensis infection in a patient with AIDS in Brazil. Clin Infect Dis. 1999;28(1):144–145. doi:10.1086/517180
24. Arends JE, Stemerding AM, Vorst SP, de Neeling AJ, Weersink AJ. First report of a brain abscess caused by Nocardia veterana. J Clin Microbiol. 2011;49(12):4364–4365. doi:10.1128/JCM.01062-11
25. Hu N, Zou W, Cai Q, et al. The first report of cerebral nocardiosis caused by nocardia terpenica together with exiguobacterium profundum bacteremia. Case report. Jundishapur J Microbiol. 2018;11(10):e69604. doi:10.5812/jjm.69604
26. Hoshino Y, Watanabe K, Iida S, et al. Nocardia terpenica sp. nov., isolated from Japanese patients with nocardiosis. Int J Syst Evol Microbiol. 2007;57(Pt7):1456–1460. doi:10.1099/ijs.0.64695-0
27. Gonzalez-Carrillo C, Millan-Sauceda C, Lozano-Garza HG, et al. Genomic changes associated with the loss of nocardia brasiliensis virulence in mice after 200 in vitro passages. Infect Immun. 2016;84(9):2595–2606. doi:10.1128/IAI.00329-16
28. Beaman BL, Black CM, Doughty F, Beaman L. Role of superoxide dismutase and catalase as determinants of pathogenicity of Nocardia asteroides: importance in resistance to microbicidal activities of human polymorphonuclear neutrophils. Infect Immun. 1985;47(1):135–141. doi:10.1128/iai.47.1.135-141.1985
29. Mannle D, McKinnie SMK, Mantri SS, et al. Comparative genomics and metabolomics in the genus nocardia. mSystems. 2020;5(3). doi:10.1128/mSystems.00125-20
30. Nouioui I, Cortes-Albayay C, Neumann-Schaal M, et al. Genomic virulence features of two novel species Nocardia barduliensis sp. nov. and Nocardia gipuzkoensis sp. nov., isolated from patients with chronic pulmonary diseases. Microorganisms. 2020;8(10):1517. doi:10.3390/microorganisms8101517
31. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi:10.1038/nmeth.1923
32. Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi:10.1093/bioinformatics/btr509
33. Arndt D, Grant JR, Marcu A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–W21. doi:10.1093/nar/gkw387
34. Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi:10.1093/bioinformatics/btv421
35. Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019;47(D1):D687–D692. doi:10.1093/nar/gky1080
36. Margalit I, Muhsen K, Ben Ari Y, et al. Nocardia colonization in contrast to nocardiosis: a comparison of patients’ clinical characteristics. Eur J Clin Microbiol Infect Dis. 2020;39(4):759–763. doi:10.1007/s10096-019-03796-5
37. Martinez Tomas R, Menendez Villanueva R, Reyes Calzada S, et al. Pulmonary nocardiosis: risk factors and outcomes. Respirology. 2007;12(3):394–400. doi:10.1111/j.1440-1843.2007.01078.x
38. Takiguchi Y, Ishizaki S, Kobayashi T, et al. Pulmonary nocardiosis: a clinical analysis of 30 cases. Intern Med. 2017;56(12):1485–1490. doi:10.2169/internalmedicine.56.8163
39. Komaki H, Ichikawa N, Hosoyama A, et al. Genome based analysis of type-I polyketide synthase and nonribosomal peptide synthetase gene clusters in seven strains of five representative Nocardia species. BMC Genomics. 2014;15:323. doi:10.1186/1471-2164-15-323
40. Nouioui I, Ha SM, Baek I, Chun J, Goodfellow M. Genome insights into the pharmaceutical and plant growth promoting features of the novel species Nocardia alni sp. nov. BMC Genomics. 2022;23(1):70. doi:10.1186/s12864-021-08257-y
41. Patra P, Mondal N, Patra BC, Bhattacharya M. Epitope-based vaccine designing of nocardia asteroides targeting the virulence factor mce-family protein by immunoinformatics approach. Int J Pept Res Ther. 2020;26(2):1165–1176. doi:10.1007/s10989-019-09921-4
42. Xu S, Wei M, Li G, et al. Comprehensive analysis of the nocardia cyriacigeorgica complex reveals five species-level clades with different evolutionary and pathogenicity characteristics. mSystems. 2022;7(3):e0140621. doi:10.1128/msystems.01406-21
43. Fatahi-Bafghi M. Nocardiosis from 1888 to 2017. Microb Pathog. 2018;114:369–384. doi:10.1016/j.micpath.2017.11.012
44. Parish T, Hatfull GF, Jacobs WR. Two-component regulatory systems of mycobacteria. Microbiol Spectr. 2014;2(1). doi:10.1128/microbiolspec.MGM2-0010-2013
45. Rifat D, Bishai WR, Karakousis PC. Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J Infect Dis. 2009;200(7):1126–1135. doi:10.1086/605700
46. Parish T, Smith DA, Roberts G, Betts J, Stoker NG. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology. 2003;149(Pt 6):1423–1435. doi:10.1099/mic.0.26245-0
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.