Back to Journals » Journal of Multidisciplinary Healthcare » Volume 16
A Comparison of Single and Combined Schemes of Asia-Pacific Colorectal Screening, Faecal Immunochemical and Stool Deoxyribonucleic Acid Testing for Community Colorectal Cancer Screening
Authors Ze Y, Tu H, Zhang L, Bai Y, Ren Y, Chen X, Xue Y, Sun R, Yang Y, Yang J, Zhou X, Liu L
Received 5 December 2022
Accepted for publication 13 February 2023
Published 1 March 2023 Volume 2023:16 Pages 571—586
DOI https://doi.org/10.2147/JMDH.S398997
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Yuan Ze,1 Huiming Tu,2 Lin Zhang,3,4 Yu Bai,5 Yilin Ren,2 Xin Chen,1 Yuzheng Xue,2 Renjuan Sun,6 Yuling Yang,7 Jie Yang,2 Xuan Zhou,2 Li Liu8
1Wuxi School of Medicine, Jiangnan University, Wuxi, People’s Republic of China; 2Department of Gastroenterology, Affiliated Hospital of Jiangnan University, Wuxi, People’s Republic of China; 3Institute of Artificial Intelligence, Hefei Comprehensive National Science Center, Hefei, People’s Republic of China; 4School of Population Medicine and Public Health, Peking Union Medical College/Chinese Academy of Medical Sciences, Beijing, People’s Republic of China; 5Department of Gastroenterology, Changhai Hospital of Shanghai, Shanghai, People’s Republic of China; 6Outpatient Nursing department, Affiliated Hospital of Jiangnan University, Wuxi, People’s Republic of China; 7Nursing department of Geriatrics Ward, Affiliated Hospital of Jiangnan University, Wuxi, People’s Republic of China; 8Data Center, Affiliated Hospital of Jiangnan University, Wuxi, People’s Republic of China
Correspondence: Huiming Tu, Department of Gastroenterology, Affiliated Hospital of Jiangnan University, No. 1000, Hefeng Road, Binhu District, Wuxi, 214122, People’s Republic of China, Tel +86-13861753621, Email [email protected]
Objective: To compare the screening efficacy of colonoscopy and pathologically confirmed single and combined Asia-Pacific colorectal screening (APCS), faecal immunochemical testing (FIT) and stool deoxyribonucleic acid (sDNA) testing protocols.
Methods: From April 2021 to April 2022, 842 volunteers participated in primary colorectal cancer (CRC) screenings using APCS scoring, FIT and sDNA testing and 115 underwent a colonoscopy. One hundred high-risk participants were then identified from the results of both processes. The differences in the three CRC screening tests in combination with the colonoscopy pathology diagnostics were evaluated using Cochran’s Q test, the Dunn–Bonferroni test and area under the receiver operating characteristic curve (AUC) value analysis.
Results: Both FIT and sDNA testing demonstrated a 100% performance in detecting CRC. For advanced adenoma, the sensitivity of the FIT + sDNA test scheme (double positive) was 29.2%, and the sensitivities of the combined FIT + sDNA test and APCS scoring + sDNA test schemes were 62.5% and 95.8%, respectively. The FIT + sDNA testing kappa value of advanced colorectal neoplasia was 0.344 (p = 0.011). The sensitivity for nonadvanced adenoma of the APCS score + sDNA test scheme was 91.1%. In terms of positive results, the sensitivity of the APCS score + FIT + sDNA detection protocol was significantly higher than that of the APCS score, FIT, sDNA detection, and FIT + sDNA detection methods (adjusted p < 0.001, respectively). For the FIT + sDNA test, the kappa value was 0.220 (p = 0.015) and the AUC was 0.634 (p = 0.037). The specificity of the FIT + sDNA test scheme was 69.0%.
Conclusion: The FIT + sDNA test scheme demonstrated superior diagnostic efficacy, and the combined APCS score + FIT + sDNA test scheme demonstrated remarkable improvements in CRC screening efficiency and sensitivity for detecting positive lesions.
Keywords: faecal immunochemical testing, stool DNA test, colonoscopy, primary screening, colorectal cancer
Novelty and Impact
The Asia-Pacific colorectal screening (APCS) score, faecal immunochemical test (FIT) and stool DNA (sDNA) test have been widely applied in colorectal cancer (CRC) screening. However, the diagnostic performances of all three in various combinations have not thus far been compared in a single study of CRC screening. As such, the effectiveness of a variety of screening schemes used to generate colonoscopy and pathology results were analysed, confirming that the combined FIT + sDNA test scheme exhibited superior diagnostic efficacy and that the combined APCS scoring + FIT + sDNA test scheme demonstrated a remarkable improvement in efficiency and the sensitivity of positive lesion results.
What is Already Known on This Topic – Summarise the State of Scientific Knowledge on This Subject Before You Did Your Study and Why This Study Needed to be Done
Non-invasive screening for CRC is effective and can reduce the incidence and mortality rates; however, the diagnostic performances of the APCS, FIT and sDNA tests in various combinations have not been compared in any individual study pertaining to CRC screening.
What This Study Adds – Summarise What We Now Know as a Result of This Study That We Did Not Know Before
The FIT + sDNA test scheme demonstrated superior diagnostic efficacy, and the combined APCS score + FIT + sDNA test scheme demonstrated remarkable improvement in terms of efficiency and sensitivity for detecting positive lesions in CRC screening.
How This Study Might Affect Research, Practice or Policy – Summarise the Implications of This Study
The primary screening programme adopted in this study using the APCS score with FIT + sDNA testing could be applied to improve screening efficiency and the diagnostic sensitivity of positive lesions.
Abbreviations
Confidence interval (CI); advanced adenoma (AA); advanced colorectal neoplasia (ACN); Asia-Pacific colorectal screening (APCS); area under the receiver operating characteristic curve (AUC); colorectal cancer (CRC); cycling threshold (CT); faecal immunochemical testing (FIT); high-grade intraepithelial neoplasia (HGIN); low-grade intraepithelial neoplasia (LGIN); multitarget stool DNA (mtsDNA); nonadvanced adenoma (NAA); receiver operating characteristic (ROC); syndecan-2 (SDC2); stool DNA (sDNA); standard error (SE).
Introduction
Colorectal cancer (CRC) is the third most common cancer worldwide, with the second highest mortality rate, presenting a serious burden on society.1 A prognosis of CRC survival is associated with the nascent stages of the disease, where only precursor lesions, such as adenomatous polyps or sessile serrated lesions, are present. The development of cancer within approximately 10 years provides an important time window for early lesion detection in screening,2,3 which has been proven to reduce CRC incidence and mortality in randomised controlled clinical trials.3,4 Currently, the methods for CRC screening differ worldwide based on population risk and resources.3 Colonoscopy with high specificity is a diagnostic criteria for CRC, and this is key to the early detection of lesions and improving the patient’s quality of life. However, due to its invasiveness, complex preparation process, patient discomfort and high cost, compliance is low.5–7
Studies have reported that a CRC screening model combining faecal immunochemical testing (FIT) and Asia-Pacific colorectal screening (APCS) scoring predicted the risk of advanced colorectal tumours in the asymptomatic Asian population,8,9 with the advantage of a convenient form resulting in good compliance. In fact, APCS scoring has achieved optimal cost-effectiveness and primary screening efficiency.8 Nevertheless, due to its characteristics of higher sensitivity and lower specificity, APCS scoring increases the burden on resources. Meanwhile, FIT is highly sensitive in detecting blood haemoglobin in the faecal lower digestive tract (70–88%) and specificity (90–96%) in CRC screening10 and has become the preferred method.11,12 Furthermore, the stool deoxyribonucleic acid (sDNA) test for DNA isolation of faecal colonic epithelial tumour cells detached from the intestinal lumen identifies mutant forms of Kirsten rat sarcoma and aberrant methylation of certain genes.13
In 2016, sDNA testing was first recognised as a screening strategy by the US Preventive Services Task Force.14 A new sDNA test targeting methylated syndecan-2 (SDC2), known as fibroglycan, encodes a transmembrane (type I) heparan sulphate proteoglycan.15 Research has confirmed that FIT improves the uptake and that sDNA testing of methylated SDC2 has an excellent diagnostic consistency with colonoscopy.13 Prior to testing, both procedures involve a period of abstinence from food and medications, as well as certain environmental restrictions.11,16 Therefore, the advantages of simplicity, high stability and non-invasive surgery offered by methods such as FIT and sDNA testing make these options effective for CRC screening.9,17
However, the reported results vary among the studies. According to the present study, the CRC sensitivity of sDNA testing is 92%, superior to that of FIT, which produced a value of 74%.18 Meanwhile, Sharma demonstrated that FIT is better than sDNA testing in reducing CRC incidence and mortality,19 while Jin et al concluded that two sDNA tests represent no significant advantage over FIT.20
Furthermore, there exist geographical discrepancies in economic resources, healthcare structure and infrastructure in nations such as China, and a large number of false-positive screening results have led to increased demand for colonoscopies.3 Thus, choosing the feasibility and reliability of a specific CRC screening method remains a significant challenge. The diagnostic performances of APCS scoring, FIT and sDNA testing in various combinations have not previously been compared in an individual study of CRC screening. As such, this study aimed to compare the screening effectiveness of single and combined APCS scoring, FIT and sDNA testing schemes as confirmed using colonoscopy and pathology.
Materials and Methods
Study Design and Participants
From 2021 to 2022, the Affiliated Hospital of Jiangnan University, Wuxi, recruited local permanent residents to participate in CRC screening. In accordance with the programme design, the inclusion criteria for the participants were as follows: ① asymptomatic (without notable symptoms or signs of CRC, such as blood in the stool, black stool, unexplained anaemia or weight loss, abdominal mass or positive digital rectal examination); ② aged 45 to 75 years; and ③ individuals willing to voluntarily participate. The exclusion criteria for participation were as follows: ① contraindication of intestinal preparation or colonoscopy; ② resected adenomas or serrated polyps (but no proliferative, inflammatory or polypoid tumours, etc.); ③ patients who have undergone a colonoscopy within the last five years; ④ patients with an inflammatory bowel disease; ⑤ patients strongly suspected of having CRC, known to have a history of rectal CRC or who had undergone imaging and laboratory tests; ⑥ patients with hereditary CRC syndrome (including polyps); ⑦ patients with organ dysfunction, such as pregnant or severe cardiopulmonary kidney; ⑧ patients with abnormal coagulability or who had taken antiplatelets or anticoagulants within the last seven days; and ⑨ individuals unwilling to participate.
All FIT and sDNA test sampling devices were provided free of charge by the project group. The participants used their mobile phones to fill in the application form on site and were given FIT and sDNA test sampling devices simultaneously. Faecal samples from the 734 participants were successfully obtained and sent to the laboratory for unified testing. Those with high-risk or positive results in the APCS scoring, FIT or sDNA testing were categorised as high-risk for CRC and were asked to proceed with a colonoscopy. All colonoscopies were carried out in a designated endoscopy centre by trained clinical research coordinators who inputted the relevant information into the system and submitted the related inspection.
In this study, a total of 100 participants underwent CRC screening and a subsequent colonoscopy. To improve compliance among the high-risk population for the primary screening and to encourage follow-up compliance, these participants were contacted by phone and were sent a brief text message, while a green channel was provided, an expert outpatient service was arranged and a colonoscopy was completed within one week of receiving the test results. According to the protocol for the blind study method, the laboratory investigator was not informed of the participants’ APCS scores, and the colonoscopy doctors were unaware of their primary screening results. The participants voluntarily offered their consent. The study followed the Declaration of Helsinki and was approved by the Ethics Committee of the Affiliated Hospital of Jiangnan University (Ethics approval number: LS2021013).
Screening Methods
Asia-Pacific Colorectal Screening
The questionnaire for the APCS score was integrated into the mobile application, and the study coordinators instructed the participants on how to complete the form on site.8 The questionnaire included information on age, gender, family history of CRC and smoking history.8 The weighting of the risk scoring system was as follows: age (<50 years: 0; 50–69 years: 2; ≥70 years: 3), gender (female: 0; male: 1), family history of CRC (absent: 0; first-degree relative: 2) and smoking history (never: 0; current or past: 1) (Supplemental Information Table S1). According to the total score, three tiers of risk were defined: 0–1 = “average risk”; 2–3 = “moderate risk” and 4–7 = “high risk”.8 As noted, those returning a high-risk score were recommended to undergo a colonoscopy.8
Faecal Immunochemical Testing
In brief, FIT technology uses antibodies to detect the globin portion of human haemoglobin concentrations and has high sensitivity and specificity in CRC screening.11,21 According to the thresholds provided by the manufacturer (Shenzhen Wodehealth Biotech Co., Ltd.), a result of >100 ng/mL is defined as positive, and as negative in all other cases.
Stool Deoxyribonucleic Acid Testing
The sDNA test kit used in this study was based on the methylated human SDC2 gene assay provided by Creative Biosciences Co., Ltd. (Guangzhou, China). In strict accordance with the product’s instructions, stool samples (approximately 4.5 g) were collected by filling the cavity at the front of a sampling syringe before this was deposited into a storage tube prefilled with 16 mL of preservative buffer. The optimum liquid level was 20 mL and could not exceed 25 mL. There were no dietary restrictions imposed prior to sampling, and no samples were collected if the participant had watery diarrhoea. Notably, the cycling threshold (CT) value ≤38 was positive, while the 38 < CT value ≤39 was negative, indicating gene mutation associated with mild intestinal cancer, and a further colonoscopy was thus recommended. However, if the CT value was >39, or there was no negative CT value, there was no mutation gene.
Sample Collection
The staff distributed sampling devices among the community along with an instructional video on how the participants should take faeces samples, which they were required to send to the staff within 24 h. After receiving the samples, the collection staff checked the quality on site to ensure that the samples met the strict requirements (eg no leakage and sufficient capacity) before finally registering the information. If the requirements were not met, the staff contacted the relevant participant to obtain a new sample; however, this was only permitted once. Following this, under normal room temperature, the approved FIT and sDNA test samples were sent to the Creative Biosciences Co., Ltd. (Guangzhou, China) laboratory on the same day. Subsequently, unified testing reports were issued by the professional experimenter within three days.
Colonoscopy and Histopathologic Examination
After issuing the primary screening results, the staff notified the participants, who were clearly informed of the benefits and risks of colonoscopy before reserving a place for surgery. All colonoscopies were operated by senior endoscopists (operation experience ≥5 years; >3000 cases) to ensure the quality of the procedure (withdrawal time ≥6 minutes; Boston Bowel Preparation Scale ≥6 and each segment ≥2). The camera was successfully inserted into the caecum of all participants. The sarcoma tissue sections were then examined by experienced pathologists. The bowel procedure carried out prior to the colonoscopy followed the guidelines provided by the Digestive Endoscopy Special Committee of Endoscopic Physicians Branch of Chinese Medical Association (2019).22 This high-quality preparation of the intestine was commenced three days before the inspection and was not permitted on the inspection day itself. In addition, intestinal drugs were used, which consisted of three boxes of compound polyethylene glycol electrolyte powder (I) (H20020031) manufactured by the Jiangxi Hengkang Pharmaceutical Co., Ltd. and one box of dimethyl silicone oil powder (H51023869) manufactured by Zigong Honghe pharmaceutical Co., Ltd.
Outcomes Confirmation
The main indexes adopted in this study were used to calculate the sensitivity and specificity within 95% confidence intervals (CIs). The sensitivity was denoted by the proportion of positive test results in each lesion group, while the specificity was the proportion of negative test results in the negative results group. Furthermore, the kappa value was used to evaluate the diagnostic consistency between each primary screening protocol and the colonoscopy pathology results. When the kappa value was <0, this indicated that there was no consistency, while when the value was >0, this indicated that the consistency was meaningful, with increased values representing increased consistency. Combined with the colonoscopy and pathological results, the most severe pathology results served as the final classification criteria.
In this study, the classification of the colonoscopy pathological findings were as follows: positive results included CRC, advanced adenoma (AA) and nonadvanced adenoma (NAA); negative results included chronic colitis, inflammatory polyps, proliferative polyps and no abnormalities. Specifically, AA was diagnosed when the adenomas (including sessile serrated lesions) exhibited diameters of >10 mm, when the adenomas had villous structures, or when high-grade intraepithelial neoplasia (HGIN) was present.13 Advanced colorectal neoplasia (ACN) included CRC and AA.23
Statistical Analysis
Due to the small sample size of this study, a nonparametric test was adopted. The characteristics of the study population were compared using the Pearson Chi-square test and the Mann–Whitney U-Test. The sensitivity and specificity were calculated using Cochran’s Q test and the exact Cochran’s Q test for CRC. Pairwise comparisons were generated using the Dunn–Bonferroni test, with the rate calculated using the 95% CI of the Clopper–Pearson test. The area under the curve (AUC) of the receiver operating characteristic (ROC) curves with 95% CIs were used to evaluate the diagnostic performance of the test. And the ROC as the negative results group as the control group. The data analysis was performed using SPSS statistical software (vers. 25.0, SPSS Inc., Chicago, Illinois, USA). All tests were two-sided, and a p-value of <0.05 or adjusted values were considered to be statistically significant.
Results
Study Population
A total of 824 participants were recruited from local communities between April 2021 and April 2022. Of these, 734 completed the primary screening, 115 (55.0%) from the CRC high-risk population of 209 (28.5%) completed the colonoscopy, and 100 participants in total from the same high-risk group underwent both the screening and a colonoscopy and were included in the final study analysis (Figure 1). According to the statistics, of the 100 participants, 63 were male (63.0%) and 37 were female (37.0%), with a median age ranging from 53 to 67 years. There was no significant difference in the basic data of the positive and negative results groups (p > 0.05), which were thus comparable (Table 1). The observations of the study population were clearly diagnosed via colonoscopy and pathology, with two (2%) cases of CRC, 24 (24%) cases of AA, 45 (45%) cases of NAA and 29 (29%) cases in the negative results group.
 |
Table 1 Characteristics of the Study Population at Baseline Stratified by the Lesion Groups |
 |
Figure 1 Flowchart of the participants. Abbreviations: FIT, fecal immunochemical test; sDNA, stool DNA. |
Analysis of the Single and Combined Screening Schemes
Of the population included in this study, at least one screening scheme resulted in each participant testing as either high risk or positive. Therefore, the sensitivity from the combined APCS score + FIT + sDNA test scheme for each lesion was 100% (Table 2). Independently, the FIT and sDNA test schemes each detected CRC with a 100% accuracy (Supplemental Information Figure S1), and the three methods were all positive for HGIN (Supplemental Information Figure S2). In terms of AA, the combined APCS score + FIT + sDNA test scheme demonstrated a significantly higher sensitivity than the FIT and sDNA test schemes (adjusted p < 0.001 and p = 0.002, respectively) (Figure 2, Supplemental Information Figure S3), while the combined APCS score + sDNA test scheme demonstrated a sensitivity of 95.8% (95% CI: 78.9–99.9) (Supplemental Information Figure S4). Furthermore, the combined FIT + sDNA test scheme exhibited a kappa value for ACN of 0.344 (95% CI: 0.095–0.593) (Table 3). In terms of NAA, the specificity of the combined APCS score + FIT + sDNA test scheme was compared to the combined APCS score, FIT, sDNA, and FIT + sDNA test scheme, and a significant difference was recorded (adjusted p = 0.009, p < 0.001, p < 0.001 and p < 0.001). The sensitivity of the combined APCS score + sDNA test scheme was significantly higher than that of the combined FIT + sDNA test scheme (91.1% vs 53.3%, adjusted p = 0.009). Furthermore, there was no statistically significant difference in the kappa value for NAA and low-grade intraepithelial neoplasia (LGIN) (0.208 vs 0.189) in the combined FIT + sDNA test scheme.
 |
Table 2 The Sensitivity and Specificity of the Single and Combined Complementary Primary Screening Schemes |
 |
Table 3 The Kappa Value of the Single and Combined Complementary Primary Screening Schemes |
In terms of the results where all the lesions were positive, the sensitivity of the combined APCS score + FIT + sDNA test scheme was significantly higher than that of the APCS score, FIT, sDNA test and combined FIT + sDNA test scheme (adjusted p < 0.001). Moreover, the sensitivity of the APCS score was 63.4% (95% CI: 51.1–74.5), and the sensitivity for the combined APCS score + sDNA test scheme was 93.0% (95% CI: 84.3–97.7), which was significantly higher than that of the FIT + sDNA test scheme at 57.7% (95% CI: 45.4–69.4, adjusted p < 0.001). In addition, the kappa value for the combined FIT + sDNA test scheme was 0.220 (95% CI: 0.046–0.394, p = 0.015) (Table 3). Following the protocol for primary screening of high-risk people, there were no negative results in the combined APCS score + FIT + sDNA test scheme, excluding comparisons. The specificity of FIT (89.7%, 95% CI: 72.6–97.8) was significantly higher than that of the APCS score, the APCS score + FIT and the APCS score + sDNA testing (adjusted p < 0.001). The specificity of the combined FIT + sDNA testing scheme was 69.0% (95% CI: 49.2–84.7) in the combined protocols, which was significantly higher than the APCS score, APCS score + FIT, and APCS score + sDNA test (adjusted p = 0.023, p < 0.001, p < 0.001, respectively), with the difference statistically significant.
Double Positive Value
When the results of all the combined schemes were positive, the sensitivity of the combined FIT + sDNA test scheme was 29.2% (95% CI: 12.6–51.1) for AA (Table 4). The sensitivity of the combined APCS score + sDNA test scheme for NAA was 13.3% (95% CI: 5.1–26.8), which was significantly higher than that of both the FIT + sDNA test and the combined APCS score + FIT + sDNA test schemes (adjusted p = 0.023) (Figure 3). For all positive lesion results, the sensitivity of the combined APCS score + sDNA test scheme was 16.9% (95% CI: 9.0–27.7).
 |
Table 4 The Double Positive Detection of the Combined Primary Screening Schemes |
Receiver Operating Characteristic Validation
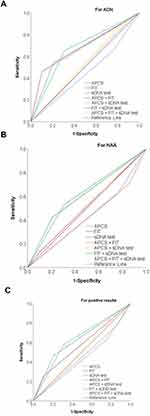
To further analyse the diagnostic value of each primary screening programme for all positive lesion results, the negative (ie non-lesion) results were used as the control group (Figure 4). In terms of ACN, the AUC of the FIT was 0.679 (95% CI: 0.534–0.824), and the AUC of the combined FIT + sDNA test scheme was 0.672 (95% CI: 0.527–0.817) (Supplemental Information Table S2). In terms of NAA, the AUC of the combined FIT + sDNA test scheme was 0.611 (95% CI: 0.480–0.743), with no significant difference (Supplemental Information Table S3). For all positive lesion results, the AUC of the combined FIT + sDNA test scheme was 0.634 (95% CI: 0.514–0.753, p = 0.037) (Supplemental Information Table S4).
Discussion
This study found that 43% of the participants had LGIN in the high-risk population and that both the APCS score + FIT and the APCS score + sDNA test schemes were effective in improving sensitivity. In addition, the combined FIT + sDNA test scheme demonstrated superior diagnostic efficacy, and the combined APCS score + FIT + sDNA test scheme further improved the sensitivity to lesions and the screening efficiency.
In previous multicentre clinical trial screening research, the sensitivity of sDNA testing for CRC ranged from 71.56% to 90.0%, while the specificity ranged from 88.2% to 100%, indicating a good diagnostic effect.13,15,24–26 However, SDC2 had an overall sensitivity of 100% and a specificity of 79.3% for CRC in this study, which may be related to the small CRC sample size. With regard to AA and ACN, the consistency of the colonoscopy findings of the present study was poor in comparison to a previous study.13 The present study also revealed that sDNA has better sensitivity than FIT for detecting LGIN, AA, and ACN, with an overall sensitivity of FIT for AA of 41.7%, corresponding to the previously reported range of 27.0%–67.0%, while the FIT specificity was higher than that of the sDNA method,27 which was consistent with the results of a previous study.20 While the FIT and sDNA test results presented some fluctuation,17 it can be stated that the risk of malignant transformation could be reduced by early lesion detection, and further colonoscopy is required to detect high-risk groups.28
On combining the low specificity of the sDNA test and the high sensitivity of FIT in this study, the attendant analysis revealed that when compared to the combined scheme, where both were positive, the scenario where either one was positive improved the overall sensitivity and made up for the missed diagnosis of double positives, thus demonstrating complementary advantages. A previous study29 reported that the combination of FIT and sDNA tests had a sensitivity of 81.5% for CRC and was diagnostically superior to single FIT and sDNA tests. Similarly, in terms of AA, the same combination had a sensitivity level of 27.8%, which was superior to that of FIT (11.1%), while the specificity was 94.4%, higher than both sDNA tests (88.9–66.7%).
Another important indicator of evaluation is specificity, which denotes the ability of a non-invasive screening technique to confirm a negative result and thus reduce the need for a follow-up colonoscopy. In the protocol adopted in this study, the FIT exhibited the highest specificity. Moreover, the specificity of the FIT + sDNA test combination was significantly better than the other pairwise combination schemes (adjusted p = 0.001), but was slightly lower than that of the single FIT and sDNA test schemes, while the difference was not significant. Consequently, when the results were negative in the single FIT and sDNA test schemes and the combined FIT + sDNA test scheme, the credibility levels were all higher.
With regard to the consistency for detecting lesions in this study, that of the combined FIT + sDNA test scheme was higher than that of the other pairwise combined schemes. In terms of ACN, specifically, the kappa value demonstrated the highest consistency with the diagnostic results of the colonoscopy. Surprisingly, the combined FIT + sDNA test scheme for ACN revealed the best double positive value (34.6%). Therefore, when the results of both the FIT and sDNA test were positive, the detection of abnormal lesions was stronger, and the decision to follow up with a colonoscopy was more compelling. In addition, in terms of ACN, the AUC of the combined FIT + sDNA test scheme (0.672) was similar to that of the FIT (0.679). For all positive lesion results and NAA detection, the combined FIT + sDNA test scheme demonstrated the best diagnostic efficiency, indicating that it performed excellently in distinguishing between the presence or absence of adenomas polyps.
Apart from in terms of CRC and HGIN, the sensitivity of the APCS score + sDNA test scheme was higher than the other schemes regarding lesion detection. Taking advantage of the highest sensitivity level for the APCS score in the single scheme fully complemented the false-negative results of the single FIT and sDNA test schemes, compensating for the insensitivity of these methods.30 Nevertheless, among the combined schemes, the APCS score + sDNA test performed significantly better than the FIT + sDNA test for NAA (adjusted p = 0.009), but there was no significant difference between the APCS score + sDNA test and the APCS score + FIT for ACN and NAA (adjusted p > 0.999,). The specificity of the APCS score + FIT and APCS score + sDNA test schemes was significantly lower than that of the FIT + sDNA test scheme, and the kappa value further revealed low or even no consistency with colonoscopy. Again, the advantage of the combined FIT + sDNA test scheme in terms of its diagnostic confidence was apparent.
Notably, unlike previous research, in this study, the APCS model score was modified with the addition of body mass index and diabetes as risk factors. The sensitivity of the modified APCS score combined with FIT for screening the ACN high-risk population (76.7%) was higher than for each scheme alone (36.7% and 70.0%, respectively).23,31 However, the modified risk factors were not accounted for in our questionnaire. Furthermore, there was, at the time, no capability for detecting CRC using APCS scoring in our study. In fact, two women were evaluated as being at moderate risk, and the FIT and sDNA test schemes revealed them to be positive. Therefore, it was speculated that the questionnaire was affected by objective factors and carried the potential risk of also detecting false negatives; in other words, the need for further examination could not be ruled out in cases where the questionnaire identified participants as being at average or moderate risk. Likewise, there may have been a greater risk of false positives in the positive APCS results, leading to an increased number of missed diagnoses and misdiagnoses. In addition, it must be noted that our CRC sample size was limited and more data should be collected for further validation.
Moreover, compliance is the key to determining the effectiveness of screening, and the compliance rate of colonoscopy in the high-risk population is an important quality control index in CRC screening. In this regard, it was demonstrated that the addition of APCS scoring to the combined FIT + sDNA test scheme produced primary screening results that were highly reliable and credible. Generally, not only could this scheme help to accurately identify high-risk members of the population for follow-up colonoscopy but could also reduce the number of missed diagnoses and misdiagnoses, as well as further improve the accuracy of the primary screening results. Similarly, the scheme could reduce the burden of follow-up colonoscopy in low-risk populations and could prove more effective and targeted for high-risk populations, thus increasing colonoscopy compliance. In addition, in districts where colonoscopy medical resources are limited, the combined APCS score + FIT + sDNA test scheme could effectively reduce waste and achieve a higher level of screening efficiency, as well as improve the adenoma detection rate and the quality of the screening results.
A cost–benefit study comparing FIT, multitarget sDNA (mtsDNA) testing and colonoscopy found that mtsDNA test prices were higher than those for FIT and that both FIT and colonoscopy were more cost-effective than mtsDNA testing.32 The market price for the sDNA test of methylated SDC2 used in the present study is also higher than that of FIT. As such, depending on the local conditions, the combined APCS + FIT + sDNA test scheme has potential feasibility in regions and countries with a high level of economic development. For regions and countries with a low level of economic development, the primary screening programme consisting of APCS scoring combined with either FIT or sDNA testing may be more convenient and more appropriate. To achieve good results with primary CRC screening and early treatment, this programme has certain feasibility. On the one hand, it reduces the cost of primary screening, making it more cost effective, while on the other, it raises the general awareness of screening and provides a warning to the asymptomatic population, thus prompting the high-risk population to undergo further examination.
This study involves a number of limitations. First, the cost-effectiveness of the various programmes for health economics were not compared, meaning further analysis is needed to identify cost-effective screening schemes. Second, the study was confined to an analysis of a single centre and a small sample of screening data, with a kappa value of <0.4, the consistency of which was not ideal; thus, the possibility of bias cannot be excluded. Third, the CRC sample size was small, and the FIT and sDNA test sensitivities were higher than in previous studies. In addition, it is necessary to rigorously analyse the performance of multiple centres and a large number of asymptomatic subjects in future research.
Conclusion
This study demonstrated that the FIT + sDNA detection protocol could improve the diagnostic efficiency of CRC primary screening for adenomas, with the combined APCS score + FIT + sDNA detection protocol exhibiting a significant improvement in CRC screening efficiency and sensitivity for detecting positive lesions. It is believed that the findings of this study provide valuable reference information for colorectal cancer screening.
Data Sharing Statement
As for reasonable needs, the data used in this study can be offered from the corresponding author.
Ethics Statement
This study was approved by the Ethics Committee of Affiliated Hospital of Jiangnan University (Ethical approval number: LS2021013). Participant voluntarily participated in the study and signed the consent.
Acknowledgments
We would like to appreciate the active participation of all participants and all the advice from all the researchers in our study. We are grateful to medical staff for assistance in sample collection and laboratory technicians detection of the samples and the provision of the data.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The study was funded by 2020 Taihu Talent Plan—high-end talents.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Goding Sauer A, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–164. doi:10.3322/caac.21601
2. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi:10.1016/S0140-6736(19)32319-0
3. Shaukat A, Levin TR. Current and future colorectal cancer screening strategies. Nat Rev Gastroenterol Hepatol. 2022;19(8):521–531. doi:10.1038/s41575-022-00612-y
4. Shaukat A, Mongin SJ, Geisser MS, et al. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013;369(12):1106–1114. doi:10.1056/NEJMoa1300720
5. Travis E, Ashley L, Pownall M, O’Connor DB. Barriers to flexible sigmoidoscopy colorectal cancer screening in low uptake socio-demographic groups: a systematic review. Psychooncology. 2020;29(8):1237–1247. doi:10.1002/pon.5443
6. Hoeck S, Van Roy K, Willems S. Barriers and facilitators to participate in the colorectal cancer screening programme in Flanders (Belgium): a focus group study. Acta Clin Belg. 2022;77(1):37–44. doi:10.1080/17843286.2020.1783906
7. Luque JS, Vargas M, Wallace K, et al. Engaging the community on colorectal cancer screening education: focus group discussions among African Americans. J Cancer Educ. 2022;37(2):251–262. doi:10.1007/s13187-021-02019-w
8. Yeoh KG, Ho KY, Chiu HM, et al.; Asia-Pacific Working Group on Colorectal Cancer. The Asia-Pacific Colorectal Screening score: a validated tool that stratifies risk for colorectal advanced neoplasia in asymptomatic Asian subjects. Gut. 2011;60(9):1236–1241. doi:10.1136/gut.2010.221168
9. Chen H, Lu M, Liu C, et al. Comparative evaluation of participation and diagnostic yield of colonoscopy vs fecal immunochemical test vs risk-adapted screening in colorectal cancer screening: interim analysis of a multicenter randomized controlled trial (TARGET-C). Am J Gastroenterol. 2020;115(8):1264–1274. doi:10.14309/ajg.0000000000000624
10. Imperiale TF, Gruber RN, Stump TE, Emmett TW, Monahan PO. Performance characteristics of fecal immunochemical tests for colorectal cancer and advanced adenomatous polyps: a systematic review and meta-analysis. Ann Intern Med. 2019;170(5):319–329. doi:10.7326/M18-2390
11. Meklin J, Syrjänen K, Eskelinen M. Colorectal cancer screening with traditional and new-generation fecal immunochemical tests: a critical review of fecal occult blood tests. Anticancer Res. 2020;40(2):575–581. doi:10.21873/anticanres.13987
12. van Rossum LG, van Rijn AF, Laheij RJ, et al. Random comparison of guaiac and immunochemical fecal occult blood tests for colorectal cancer in a screening population. Gastroenterology. 2008;135(1):82–90. doi:10.1053/j.gastro.2008.03.040
13. Wang J, Liu S, Wang H, et al. Robust performance of a novel stool DNA test of methylated SDC2 for colorectal cancer detection: a multicenter clinical study. Clin Epigenetics. 2020;12(1):162. doi:10.1186/s13148-020-00954-x
14. Bibbins-Domingo K, Grossman DC, Curry SJ, et al.; US Preventive Services Task Force. Screening for colorectal cancer: US preventive services task force recommendation statement. JAMA. 2016;315(23):2564–2575. doi:10.1001/jama.2016.5989
15. Niu F, Wen J, Fu X, et al. Stool DNA test of methylated syndecan-2 for the early detection of colorectal neoplasia. Cancer Epidemiol Biomarkers Prev. 2017;26(9):1411–1419. doi:10.1158/1055-9965.EPI-17-0153
16. Ransohoff DF, Lang CA. Screening for colorectal cancer with the fecal occult blood test: a background paper. American College of Physicians. Ann Intern Med. 1997;126(10):811–822. doi:10.7326/0003-4819-126-10-199705150-00014
17. Singal AG, Gupta S, Skinner CS, et al. Effect of colonoscopy outreach vs fecal immunochemical test outreach on colorectal cancer screening completion: a randomized clinical trial. JAMA. 2017;318(9):806–815. doi:10.1001/jama.2017.11389
18. Carethers JM. Fecal DNA testing for colorectal cancer screening. Annu Rev Med. 2020;71:59–69. doi:10.1146/annurev-med-103018-123125
19. Sharma T. Analysis of the effectiveness of two noninvasive fecal tests used to screen for colorectal cancer in average-risk adults. Public Health. 2020;182:70–76. doi:10.1016/j.puhe.2020.01.021
20. Jin P, You P, Fang J, et al. Comparison of performance of two stool DNA tests and a fecal immunochemical test in detecting colorectal neoplasm: a multicenter diagnostic study. Cancer Epidemiol Biomarkers Prev. 2022;31(3):654–661. doi:10.1158/1055-9965.EPI-21-0991
21. Young GP, Symonds EL, Allison JE, et al. Advances in fecal occult blood tests: the FIT revolution. Dig Dis Sci. 2015;60(3):609–622. doi:10.1007/s10620-014-3445-3
22. Digestive Endoscopy Special Committee of Endoscopic Physicians Branch of Chinese Medical Association; Cancer Endoscopy Committee of China Anti-Cancer Association. 中国消化内镜诊疗相关肠道准备指南(2019 上海)[Chinese guideline for bowel preparation for colonoscopy (2019, Shanghai)], Zhonghua Nei Ke Za Zhi. 2019;58(7):485–495. Chinese. doi:10.3760/cma.j.issn.0578-1426.2019.07.002
23. He XX, Yuan SY, Li WB, et al. Improvement of Asia-Pacific colorectal screening score and evaluation of its use combined with fecal immunochemical test. BMC Gastroenterol. 2019;19(1):226. doi:10.1186/s12876-019-1146-2
24. Wang Z, Shang J, Zhang G, et al. Evaluating the clinical performance of a dual-target stool DNA test for colorectal cancer detection. J Mol Diagn. 2022;24(2):131–143. doi:10.1016/j.jmoldx.2021.10.012
25. Han YD, Oh TJ, Chung TH, et al. Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin Epigenetics. 2019;11(1):51. doi:10.1186/s13148-019-0642-0
26. Su WC, Kao WY, Chang TK, et al. Stool DNA test targeting methylated syndecan-2 (SDC2) as a noninvasive screening method for colorectal cancer. Biosci Rep. 2021;41(1)):BSR20201930. doi:10.1042/BSR20201930
27. Malagón M, Ramió-Pujol S, Serrano M, et al. New fecal bacterial signature for colorectal cancer screening reduces the fecal immunochemical test false-positive rate in a screening population. PLoS One. 2020;15(12):e0243158. doi:10.1371/journal.pone.0243158
28. Dong J, Ma TS, Xu YH, et al. Characteristics and potential malignancy of colorectal juvenile polyps in adults: a single-center retrospective study in China. BMC Gastroenterol. 2022;22(1):75. doi:10.1186/s12876-022-02151-x
29. Mo S, Wang H, Han L, et al. Fecal multidimensional assay for non-invasive detection of colorectal cancer: fecal immunochemical test, stool DNA mutation, methylation, and intestinal bacteria analysis. Front Oncol. 2021;11:643136. doi:10.3389/fonc.2021.643136
30. Bai Y, Liu J, Kang Q, et al. Value of the methylation status of SDC2 and SFRP2 for colorectal cancer screening. Chin J Dig Endosc. 2019;36(6):427–432. doi:10.3760/cma.j.issn.1007-5232.2019.06.009
31. Luu XQ, Lee K, Kim J, Sohn DK, Shin A, Choi KS. The classification capability of the Asia Pacific Colorectal Screening score in Korea: an analysis of the Cancer Screenee Cohort. Epidemiol Health. 2021;43:e2021069. doi:10.4178/epih.e2021069
32. Ladabaum U, Mannalithara A. Comparative effectiveness and cost effectiveness of a multitarget stool DNA test to screen for colorectal neoplasia. Gastroenterology. 2016;151(3):427–439.e6. doi:10.1053/j.gastro.2016.06.003
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.