Back to Journals » Cancer Management and Research » Volume 11
A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma
Authors Zheng H, Chen T, Li C, Xu C, Ding C, Chen J, Ju S, Zhang Z, Liang Z , Cui Z, Zhao J
Received 1 October 2018
Accepted for publication 7 November 2018
Published 3 January 2019 Volume 2019:11 Pages 443—454
DOI https://doi.org/10.2147/CMAR.S189338
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Rituraj Purohit
Huifei Zheng,1,* Tengfei Chen,2,* Chang Li,2 Chun Xu,2 Cheng Ding,2 Jun Chen,2 Sheng Ju,2 Zhiwei Zhang,2 Zhipan Liang,2 Zihan Cui,2 Jun Zhao2
1National Center for Liver Cancer, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai, China; 2Department of Thoracic and Cardiovascular Surgery, The First Affiliated Hospital of Soochow University, Soochow University, Suzhou, Jiangsu, China
*These authors contributed equally to this work
Purpose: Most recently, circular RNAs (circRNAs) were considered playing regulatory roles in tumor initiation and development. The specific function of circRNAs in hepatocellular carcinoma (HCC) remains unknown. This study was designed to detect specific roles of a circRNA hsa_circ_0079299 in HCC.
Methods: The expression of hsa_circ_0079299 in HCC and tumor cell lines was detected using quantitative PCR (qPCR). Cell proliferation, migration, cell cycle and apoptosis after overexpression of the circRNA were measured using cell counting kit-8 (CCK8) assay, colony formation, 5-ethynyl-2′-deoxyuridine (EdU) assay, wound healing assay, transwell culture system and flow cytometry. Western blotting assay detected the protein expression of PI3K/AKT/mTOR signaling pathway and cyclin B1 (CCNB1). Overexpression of the circRNA in vivo was measured by nude mice tumorigenesis.
Results: The expression of hsa_circ_0079299 was lower in HCC tissues. Overexpression of hsa_circ_0079299 suppressed tumor growth in vitro and in vivo, retarded cell cycle progression while had no effect on cell migration and apoptosis. The inhibitory effect of hsa_circ_0079299 was partly mediated by PI3K/AKT/mTOR signaling pathway.
Conclusion: Our study shows that tumor suppressive role of hsa_circ_0079299 in HCC provides new recognition of circRNAs in cancers.
Keywords: cell proliferation, cell cycle, cyclin B1, liver cancer
Introduction
Globally, hepatocellular carcinoma (HCC) is the fifth most common cancer with more than 850,000 new cases yearly. This neoplasm accounts for up to 90% of all primary liver cancer and is the second leading cause of cancer-related deaths.1,2 The prevalence of well-established risk factor hepatitis B virus (HBV) infection accounted for high rates of HCC especially in Asia and Africa.3 In developed countries, another risk factor hepatitis C virus (HCV) infection increased by injection drug abuse caused elevated HCC incidence. Aflatoxin B1, cirrhosis, obesity and type 2 diabetes as less common risk factors also contributed to disease risk.4 Lack of early symptoms and possessing a prolonged subclinical course, HCC was usually diagnosed at intermediated or advanced stages with limited curative approaches.5,6 Hepatocarcinogenesis was believed to be complex multiple processes driven by genetic alternations combined with epigenetic modifications.7,8 Given the poor strategies for diagnosis and treatment, new insight of molecular pathogenesis in HCC initiation is needed which might subsequently provide potential biomarker-driven treatments.
Circular RNA (circRNA) is a special class of noncoding RNAs which originates from back splicing or exon skipping of precursor mRNA templates. Over two decades ago, circRNAs were considered inert byproducts of linear RNA aberrant splicing. With the advent of high-throughput deep sequencing, the ubiquity and abundance of circRNAs were recognized.9–11 Recent study has shown that many circRNAs highly expressed in a cell type-specific or tissue-specific manner.12
Accumulating evidence has demonstrated that circRNAs exert functions in cancer.13,14 circRNAs profiles in three paired of HCC have been reported by microarray and hundreds of upregulated circRNAs and downregulated circRNAs were identified. However, specific roles of different circRNAs in HCC remain unclear.
In the present study, we investigate the tumor suppressive role of hsa_circ_0079299 in HCC using human paired HCC and para-cancerous tissues. hsa_circ_0079299 was overexpressed to further explore the functions and mechanisms in tumor growth.
Materials and methods
Tissues and cell lines
We collected frozen liver samples including HCC and paired adjacent liver tissues from 42 patients undergoing liver surgical procedures. Diagnosis was confirmed by histological pathology. All patients gave consents to collect tissues. The ethics committee of Eastern Hepatobiliary Surgery Hospital approved this tissue study, and written informed consent was obtained from all patients. HCC cell lines SK-Hep-1, SMMC-7721, 7702, LM-3 and PRL-RF5 were maintained in DMEM with 10% FBS or 1640 medium with 10% FBS. All cell lines were purchased from Cell Bank of Chinese Academy of Sciences (Shanghai, China).
Plasmid construction and cell transfection
The complementary DNA (cDNA) of exon 2 (total 660 bp) was cloned by primers (Table 1) from total cDNA then digested with KpnI and BamHI restriction enzymes and cloned into pCD-ciR (Geneseed, Guangzhou, China). Overexpression of hsa_circ_0079929 was verified by direct sequencing. Cells are transiently transfected with plasmids using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). The expression of hsa_circ_0079929 in transfected cells was detected by quantitative PCR (qPCR) using specific primer pairs.
  | Table 1 Primers allowed for amplification of genes in this work Abbreviations: F, forward primer; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; R, reverse primer; RT, reverse transcript. |
Total RNA isolation, RNase R treatment and real-time qPCR analysis
Total RNA was extracted from HCC tissues and cell lines using TRIzol reagent (Thermo Fisher Scientific). RNase R treatment was carried out as Panda A described previously.15 When we treated with RNase R, we divided total RNA into two groups including no RNase R treatment and RNase R treatment. The first group was as common internal control when we detected hsa_circ_0079929 expression in both groups. qPCR was carried out using the SYBR Select Master Mix kit (cat. no. 4472908; Thermo Fisher Scientific).
Cell counting kit-8 (CCK8) assay
Cells were seeded at 3×103 per well in 96-well plates after transfection and incubated for 24, 48, 72 and 96 hours, and then 10 µL of WST-8 was added to each well. A plate reader (Molecular Devices LLC, Sunnyvale, CA, USA) was used to measure the absorbance at 450 nm after 1 hour at 37°C.
Colony formation assay
Cells were seeded at 400 per well in 6-well plates after transfection and incubated for 2 weeks. Until colonies were visible with naked eyes, they were fixed with 4% paraformaldehyde for 30 minutes and stained with 0.1% crystal violet, the number of colonies was then counted.
5-ethynyl-2′-deoxyuridine (EdU) assay
Transfected cells were seeded at 8,000 per well in 96-well plates overnight. According to the manual of a EdU labeling/detection kit (Ribobio, Guangzhou, China), cells were incubated with EdU at 50 µM for 2 hours at 37°C. After EdU labeling, cells were washed with PBS and fixed with 4% paraformaldehyde for 30 minutes and incubated with 2 mg/mL glycine for 5 minutes. The cells were rinsed once with PBS of 0.5% Triton X-100 (PBS) then stained with Apollo working solution at room temperature for 30 minutes in the dark. After staining, cells were washed several times with TBS of 0.5% Triton X-100 and then stained with Hoechst at room temperature for 30 minutes in a dark place. The cells were imaged by fluorescence microscopy. The percentage of EdU-positive cells was calculated from five random fields in three wells.
Flow cytometry cell cycle analysis
Cells were seeded at 105 per well in 6-well plates after transfection, incubating plates for 24, 48, 72 and 96 hours. Cells were harvested with trypsin and washed twice with cold PBS. Then, cells were resuspended in 400 µL of propidium iodide (PI) staining solution containing 0.1 mg/mL PI and 0.02 mg/mL ribonuclease A. Samples were incubated in the dark for 30 minutes at room temperature before acquisition. Acquisition was performed on FACS Calibur flow cytometer (BD Biosciences, San Jose, CA, USA) with CellQuest software. Data analysis was performed using FlowJo software.
Wound healing assay
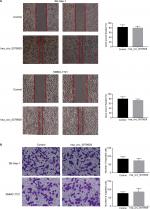
Cells were plated at 100% confluence after transfection. Then, monolayer cells were scratched with a 200 mL pipette tip across the central of the well. After rinsing detached cells with PBS one time, replenished with serum-free medium and took photos on microscope. Cells were incubated for another 24 hours and photos were taken for wound closure on the microscope. ImageJ was used to measure wound area.
Transwell invasion assay
To assess cell invasion ability, we used Matrigel (BD Biosciences) coated transwell inserts with 200 µL basal medium, which were resuspended 5×104 cells according to manufacturer’s instructions. The lower chamber contained 600 µL completed medium with 10% FBS. Cells were incubated with another 24 hours. After wiping out cells in the upper surface of the chamber, bottom cells were fixed with 4% paraformaldehyde for 15 minutes and then stained with crystal violet.
Western blotting
Total protein was extracted from transfected cells using cell lysis buffer containing 1 mM phenylmethanesulfonyl fluoride. Nuclear protein was extracted using Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime, China). Protein quantification was determined by bicinchoninic acid assay. Lysates including 30 µg protein per well were separated on 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes. Blocking with 5% nonfat milk in TBST for 1 hour was followed. After washing three times with TBST, membranes were incubated with primary antibodies and secondary antibodies in turn. Primary antibodies including p85 PI3K, p-p85 PI3K AKT, p-AKT, mTOR, p-mTOR and cyclin B1 (CCNB1) were all purchased from CST. Membranes were washed three times before detecting protein bands using enhanced chemiluminescence substrate on Tanon 5200 system.
Tumorigenicity assay
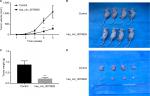
Transfected cells were digested with trypsin and washed twice with PBS. Cells were resuspended in 200 µL solution containing 100 µL PBS and 100 µL Matrigel substrate at a final density of 2×106 cells/0.2 mL. Four-week-old male BALB/c nude mice were subcutaneously injected with 2×106 tumor cells. Tumor volume was measured every 1 week. Mice were sacrificed at day 35 since tumor cells have been injected and tumor volume was measured subsequently. Tumor volume was calculated as follows: Tumor volume = length × width2/2.16 Nude mice were housed in a specific pathogen-free room. The studies of nude mice were approved by the Institutional Animal Care and Use Committee of the Soochow University, and experiments were also conducted according to the Institutional Animal Care and Use Committee of Soochow University.
Statistical analyses
Data analyses were performed using GraphPad Prism 7.0 software. We performed all experiments at least three times using transfected cells. Data are mean ± SEM. Unpaired and two-tailed Student’s t-test was used to determine statistical significance of two groups. ANOVA and post hoc two-tailed Bonferroni test was used to determine statistical significance of more than two groups. P<0.05 was considered significant. *P<0.05, **P<0.01, ***P<0.001.
Results
Downregulated hsa_circ_0079929 in liver tissue samples and liver tumor cell lines
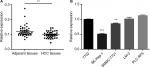
Previous research has shown that CDK13 gene was amplified and overexpressed in HCC with potential oncogenic activity.17 circRNA termed hsa_circ_0079929 generated by parent gene CDK13 has been significantly downregulated in HCC tumor tissues as compared with that of para-cancerous tissues according to circRNA expression profile by using circular microarray.18 Then, the hsa_circ_0079929 expression profile was assayed in 42 pairs of HCC tumor tissues and matched para-cancerous tissues. We confirmed the significant downregulation of hsa_circ_0079299 in tumor tissues compared with nontumoral counterparts (Figure 1A). Next, we detected the expression of hsa_circ_0079299 in liver normal cell 7702 and tumor cells including SK-HEP-1, SMMC-7721, LM3 and PLC-RF5 (Figure 1B). qPCR showed that SK-HEP-1 and SMMC-7721 expressed lower level of hsa_circ_0079299 when compared with normal liver cell line. Taken together, the expression of hsa_circ_0079299 decreased in HCC and liver tumor cell lines.
Characterization of hsa_circ_0079929 in HCC
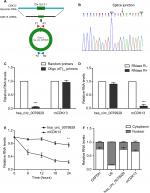
Distinguishing from those of linear counterparts, the formation of circRNA is back splicing. To be specific, hsa_circ_0079929 was generated by back-splicing exon 2 of CDK13 gene (Figure 2A). And the resulting RNA circle ligation site was located using Sanger sequencing (Figure 2B). Next, we confirmed the circularization of hsa_circ_0079929. Using random hexamer and oligo (dT)18 primers followed by qPCR, the expression of hsa_circ_0079929 was reduced significantly using oligo (dT)18 primer when compared with that of random hexamer primer (Figure 2C). This demonstrated that hsa_circ_0079929 was without the structure of poly-A tail. Furthermore, we treated RNA which has been harvested from HCC cell line SK-hep-1 with or without RNase R, a 3´ to 5´ exoribonuclease that specifically digests linear RNAs. qPCR showed hsa_circ_0079929 decreased significantly when compared with that of CDK13 mRNA (Figure 2D), the result indicated that hsa_circ_0079929 was resistant to RNase R with circular structure. Furthermore, transcriptional inhibitor actinomycin D treatment was used to measure half-life of hsa_circ_0079929 and CDK13 mRNA, we found the half-life of hsa_circ_0079929 exceeded that of linear RNA (Figure 2E), which indicated that hsa_circ_0079929 was more stable than linear RNA. Additionally, qPCR showed the circular form of CDK13 preferentially distributed in cytoplasm than in nucleus (Figure 2F). In summary, our findings showed that hsa_circ_0079929 is an abundant and stable circRNA in human cancer cells.
Overexpression of hsa_circ_0079929 suppressed tumor cell proliferation and cell cycle progression
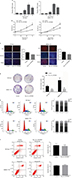
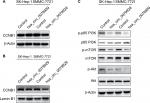
Based on the abovementioned conclusions, specific function of hsa_circ_0079929 in tumor formation and development was proposed to further study. We have confirmed the lower expression of hsa_circ_0079929 in tumor cell lines SK-HEP-1 and SMMC-7721. Next, we constructed plasmid vector of overexpressed hsa_circ_0079929, and this plasmid vector was used to transfect both cell lines. After transfection, the expression of hsa_circ_0079929 was highly increased which indicated the success of transfection and overexpression of hsa_circ_0079929 (Figure 3A). At the same time, we obtained stably overexpressing cell line using SK-HEP-1. Cell counting kit-8 (CCK8), EdU assay and colony formation revealed overexpression of hsa_circ_0079929 inhibited cell proliferation significantly compared with negative control (Figure 3B–D). What is more, we investigated the effect of hsa_circ_0079929 on cell cycle. To be specific, we measured the percentage of tumor cells in different cell cycle stages including G1/G0, S and G2/M. Overexpression of hsa_circ_0079929 increased the percentage of cells in S and G2/M while decreased the percentage of cells in G1/G0 using flow cytometry (Figure 3E). We further confirmed the expression of CCNB1 which is associated with cell cycle was downregulated in nucleus but had no change in total after overexpression of hsa_circ_0079929 (Figure 4A, B). In addition, we also detected the apoptosis of transfected cells compared with negative control, the results showed no difference (Figure 3F). In conclusion, hsa_circ_0079929 inhibited tumor cell proliferation and exerted cell cycle arrest, but had no effect on cell apoptosis.
Effects of hsa_circ_0079929 on tumor cell migration and invasion
We found that hsa_circ_0079929 significantly suppressed tumor growth, and we subsequently investigated the effect of hsa_circ_0079929 on other tumor cell functions including migration and invasion. Wound healing assay and transwell assay indicated no remarkable difference compared with negative control (Figure 5A, B). This suggested that hsa_circ_0079929 had no significant influence on cell migration and invasion.
Overexpression of hsa_circ_0079929 inhibited PI3K/AKT/mTOR signaling pathway to suppress tumor cell proliferation
To understand molecular mechanism illustrating specific function of hsa_circ_0079929 in HCC, we tested several signaling pathways during cell proliferation at protein levels by western blotting. Overexpression of hsa_circ_0079929 decreased expression of p-p85 PI3K, p-AKT and p-mTOR (Figure 4C). These results indicated that hsa_circ_0079929, at least, partially inhibited cell proliferation through PI3K/AKT/mTOR signaling pathway.
Tumorigenicity assay
To further assess whether overexpression of hsa_circ_0079929 could suppress tumor formation in vivo, we inoculated stable overexpressed hsa_circ_0079929 with SK-HEP-1 cell line subcutaneously into 4-week-old nude mice. The results showed that tumor size and weight were remarkably reduced when transplanted with tumor cells overexpressed hsa_circ_0079929 (Figure 6). Therefore, we concluded that overexpression of hsa_circ_0079929 also suppressed tumorigenesis in vivo.
Discussion
circRNAs vary in different tissues and cell types along with different roles.12 Of increasing interest is the emerging function of circRNAs in tumorigenesis. Herein, we explored the role of hsa_circ_0079929, deriving from CDK13 gene, in the pathogenesis of HCC.
In the present study, we found that hsa_circ_0079929 low expressed in HCC in comparison to matched para-cancerous samples using qPCR. Based on the downregulation of hsa_circ_0079929, we further explored its specific function and associated mechanism in HCC. We provided evidence that ectopic expression of hsa_circ_0079929 suppressed cell proliferation and retarded cell cycle progression at G2/M phase. To recognize specific function of hsa_circ_0079929 in cell proliferation, we found that cell cycle regulatory protein CCNB1 expression in total was no change while that of in nucleus was reduced. In cell cycle progression of normal cells, cyclin B1 partners with CDK1, as soon as CCNB1-CDK1 is phosphorylated and activated in the cytoplasm, it is rapidly transported into nucleus and controls cells to enter mitosis and induces G2/M transition.19–23 Prevention entry of CCNB1-CDK1 into nucleus hampers cells from initiating mitosis and subsequently accumulates in G2 phase, then finally inhibits cells from dividing into two daughter cells.24–26 To better understand the correlation between circRNAs and cell cycle progression, recent study demonstrated that circRNA circFoxo-3 combined with p21 and CDK2 composing ternary complexes, which then induced cell cycle arrest.27 In an attempt to elucidate mechanism of hsa_circ_0079929 acting as a suppressive effector on cell proliferation. We detected several canonical signaling pathways associated with cell proliferation at protein levels. We found that p85 PI3K, p-AKT and p-mTOR protein levels were decreased following hsa_circ_0079929 overexpression. Phosphatidylinositol 3-kinase (PI3K) signaling pathway as the most common pathway in cell proliferation, metabolism and apoptosis possesses downstream mediators AKT (Ser/Thr protein kinase) and mTOR (mammalian target of rapamycin).28–30 Activated p85 as subunits of phosphatidylinositol 3-kinase phosphorylates AKT, then the activated AKT enables the activation of mTOR.30 In line with our study, another CDK13-derived circRNA hsa_circ_0001699 was recently demonstrated to suppress cell proliferation, migration and invasion in HCC through JAK/STAT and PI3K/AKT pathways.31 hsa_circ_0001699 derived from four exons of CDK13 gene. Differentiating from the multiple functions of hsa_circ_0001699, our results showed limited function on cell cycle and cell proliferation but had no effects on cell migration, invasion or apoptosis, whereas regulating mechanism was in a similar way.
Given the abundant and tissue/cell-specific expression patterns of circRNAs, potential biological functions are expected. Several functions proposed for circRNA were as follows: 1) circRNAs contain binding sites for miRNA and act as sponges for miRNA, in which way could circRNAs modulate miRNAs activity.32,33 2) Regulating alternative splicing of host gene. Once circRNAs are produced, the remaining pre-mRNA transcriptions would be altered.34 3) Combining with various proteins as RNA binding protein sponges to regulate associated proteins function.27 4) circRNAs encode proteins in a cap-independent translation way.35
The hsa_circ_0079929 downstream suppressive cell proliferation and cell cycle progression effect included downregulated nuclear CCNB1. The precise regulation of hsa_circ_0079929 on CCNB1 protein was not well demonstrated in our study. Abovementioned functions of sponging miRNA or directly interacting with proteins may take predominance. Further research is needed to confirm the specific mechanism of hsa_circ_0079929 in tumor suppression.
Conclusion
Our study demonstrates low expression of hsa_circ_0079929 in HCC. Ectopic expressed hsa_circ_0079929 exerted suppressive effects on tumor cell proliferation and cell cycle progression.
Cell cycle associated protein CCNB1 and PI3K/AKT/mTOR signaling pathway were involved in the regulation of hsa_circ_0079929 functions. Despite the precise mechanism of the circRNA action is needed to be further explored, we still provided a novel insight of circRNAs as a potential therapeutic target.
Acknowledgment
This work was funded by the National Natural Science Foundation of China (Grant No. 81672934).
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. | ||
Averhoff F. Infectious Diseases Related to Travel: Hepatitis B. Centers for Disease Control and Prevention. Available from: https://wwwnc.cdc.gov/travel/yellowbook/2018/infectious-diseases-related-to-travel/hepatitis-b. Accessed December 11, 2018. | ||
Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47 Suppl:S2–S6. | ||
Bruix J, Sherman M; American Association for the Study of Liver Diseases. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53(3):1020–1022. | ||
European Association for the Study of the Liver; European Organisation for Research and Treatment of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56(4):908–943. | ||
Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. | ||
Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44(6):694–698. | ||
Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. | ||
Lu T, Cui L, Zhou Y, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21(12):2076–2087. | ||
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. | ||
Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. | ||
Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013. | ||
Chen X, Fan S, Song E. Noncoding RNAs: new players in cancers. Adv Exp Med Biol. 2016;927:1–47. | ||
Panda AC, Gorospe M. Detection and analysis of circular RNAs by RT-PCR. Bio Protoc. 2018;8(6):e2775. | ||
Naito S, von Eschenbach AC, Giavazzi R, Fidler IJ. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46(8):4109. | ||
Kim HE, Kim DG, Lee KJ, et al. Frequent amplification of CENPF, GMNN and CDK13 genes in hepatocellular carcinomas. PLoS One. 2012;7(8):e43223. | ||
Cui S, Qian Z, Chen Y, Li L, Li P, Ding H. Screening of up- and downregulation of circRNAs in HBV-related hepatocellular carcinoma by microarray. Oncol Lett. 2018;15(1):423–432. | ||
Berridge MJ. Module 9: cell cycle and proliferation. Cell Signalling Biology. 2014;6:csb0001009. | ||
Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079–3093. | ||
Xue L, Wu Z, Liu J, Luo J. FPHPB inhibits gastric tumor cell proliferation by inducing G2-M cell cycle arrest. Biomed Pharmacother. 2018;98:694–700. | ||
Gavet O, Pines J. Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and the cytoplasm at mitosis. J Cell Biol. 2010;189(2):247–259. | ||
Qi F, Chen Q, Chen H, et al. WAC promotes polo-like kinase 1 activation for timely mitotic entry. Cell Rep. 2018;24(3):546–556. | ||
Bonelli P, Tuccillo FM, Borrelli A, Schiattarella A, Buonaguro FM. CDK/CCN and CDKI alterations for cancer prognosis and therapeutic predictivity. Biomed Res Int. 2014;2014:361020. | ||
Henderson L, Bortone DS, Lim C, Zambon AC. Classic “broken cell” techniques and newer live cell methods for cell cycle assessment. Am J Physiol Cell Physiol. 2013;304(10):C927–C938. | ||
Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. | ||
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. | ||
Zarogoulidis P, Lampaki S, Turner JF, et al. mTOR pathway: a current, up-to-date mini-review (Review). Oncol Lett. 2014;8(6):2367–2370. | ||
Yu JS, Cui W. Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development. 2016;143(17):3050–3060. | ||
Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol. 2012;4(9):a011189. | ||
Lin Q, Ling YB, Chen JW, et al. Circular RNA circCDK13 suppresses cell proliferation, migration and invasion by modulating the JAK/STAT and PI3K/AKT pathways in liver cancer. Int J Oncol. 2018;53(1):246–256. | ||
Piwecka M, Glažar P, Hernandez-Miranda LR, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357(6357):eaam8526. | ||
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. | ||
Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. | ||
Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell. 2017;66(1):9–21.e7. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.