Back to Journals » OncoTargets and Therapy » Volume 13
A Circular RNA Derived from Golgi Glycoprotein 1 mRNA Regulates KRAS Expression and Promotes Colorectal Cancer Progression by Targeting microRNA-622
Authors Hao S, Qu R, Hu C, Wang M, Li Y
Received 26 September 2020
Accepted for publication 29 November 2020
Published 8 December 2020 Volume 2020:13 Pages 12637—12648
DOI https://doi.org/10.2147/OTT.S284032
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Takuya Aoki
Shuhong Hao,1 Rongfeng Qu,1 Chunmei Hu,1 Min Wang,2 Yarong Li1
1Department of Hematology and Oncology, The Second Hospital of Jilin University, Changchun, Jilin, People’s Republic of China; 2Department of General Surgery, The Second Hospital of Jilin University, Changchun, Jilin, People’s Republic of China
Correspondence: Yarong Li; Min Wang Ziqiang Street No. 265, Changchun, Jilin 130041, People’s Republic of China
Tel +86 431 81136827
; +86 431 81136427 Fax +86 431 81136827; +86 431 81136427 Email [email protected]; [email protected]
Background: Circular RNAs (circRNAs) represent a distinct class of non-coding RNAs that have attracted substantial research attention in recent years. We identified a novel circRNA derived from golgi glycoprotein 1 mRNA (circ_GLG1), the role of which is unknown in colorectal cancer (CRC). The purpose of this study was to explore the potential roles and mechanisms of circ_GLG1 in CRC.
Materials and Methods: Quantitative reverse transcriptase-polymerase chain reaction analysis was performed to quantify circ_GLG1 expression in 40 pairs of CRC tissues and adjacent normal tissues as well as CRC cell lines. DLD1 CRC cells were transfected with a small-interfering RNA against circ_GLG1, after which cell proliferation, viability, invasion, and migration were measured through cell counting kit-8 colony-formation, transwell, and wound-healing assays, respectively. Dual-luciferase reporter assays were performed to explore the binding sites among circ_GLG1, miR-622, and Kirsten rat sarcoma (KRAS) transcripts. KRAS protein expression was detected using Western blot analysis.
Results: Circ_GLG1 expression was significantly higher in CRC tissues than in adjacent normal tissues. Knocking down circ_GLG1 in DLD1 cells inhibited tumor cell viability, proliferation, invasion, and migration, and these effects were reversed by co-transfecting an miR-622 inhibitor. Circ_GLG1 promoted KRAS expression at both the mRNA and protein levels by acting as an miR-622 sponge. Dual-luciferase reporter assays demonstrated that miR-622 interacted with circ_GLG1 and KRAS mRNA.
Conclusion: Our study revealed the role of the circ_GLG1–miR-622–KRAS axis in CRC. Moreover, our findings provide insight into the molecular mechanism of circ_GLG1 in CRC and suggest potential new biomarkers for diagnosing this disease.
Keywords: colorectal cancer, circ_GLG1, miR-622, KRAS
Introduction
Colorectal cancer (CRC) is the third most common cancer in the world and seriously endangers human health.1 In the United States, 140,250 new cases of CRC were diagnosed, and 50,630 patients died of the disease in 2018.2 In China, CRC has the fifth highest prevalence rate and is the fifth leading cause of cancer-related death.3 With the development of diagnostic and treatment technologies for CRC, the prognosis of patients has improved owing to early detection. However, because the early symptoms are not evident and screening methods are limited, many patients with CRC are diagnosed at an advanced stage, which seriously affects disease prognosis.4 Therefore, there is an urgent need to explore other unknown mechanisms that contribute to CRC tumorigenesis and identify biomarkers for early diagnosis and therapy.
Circular RNAs (circRNAs) constitute a distinct type of endogenous non-coding RNA molecules with a closed-loop structure.5 CircRNAs were first discovered in 1976 and initially considered to be by-products of precursor mRNAs; therefore, they did not attract substantial attention.6 However, with the development of high-throughput sequencing, researchers have identified thousands of circRNAs in viruses, fungi, plants, animals, and other organisms.7 Further research has shown that circRNAs regulate gene expression at the epigenetic, transcriptional, and post-transcriptional levels.8 Recent findings have shown that circRNAs are abnormally expressed in various types of tumors (including CRC) and can act as oncogenes or tumor-suppressor genes by helping regulate the proliferation, invasion, migration, and apoptosis of tumor cells, thereby affecting tumorigenesis and development; examples include CiRS-7 in non-small cell lung cancer,9 circSLC26A4 in cervical cancer,10 circABCB10 in liver cancer,11 hsa-circ-0006984 in esophageal cancer,12 and hsa-circ-0003221 in bladder cancer.13 Unlike linear RNAs, circRNAs do not have a 5′ cap or 3′ polyadenylated tail structure and instead form a covalently closed ring structure, which renders them resistant to digestion via exonucleases such as RNases and confers high stability.14 In addition, circRNAs are highly conserved and serve specific roles in different species and tissues.15 CircRNAs are stably expressed in tissues, blood, sputum, and exosomes and can be detected using simpler methods than those used to detect proteins.16 Based on these characteristics, circRNAs are expected to have the greatest potential as biomarkers for tumor diagnosis and prognosis. At present, the most important known functions of circRNAs are as microRNA (miRNA) sponges, which enhance the expression levels of miRNA-target genes by adsorbing miRNA molecules.17,18 For example, Han et al found that circBANP can promote lung cancer progression by regulating miR-503 and LARP1 expression.19 Zhong et al revealed that the circMYLK–miR-29a–VEGFA-interaction network plays important roles in the proliferation and invasion of bladder cancer.20 Although in recent years, great progress has been made in the study of circRNA in cancers, the exact functions and mechanisms of circRNAs in cancers, especially CRC, remain unclear.
By performing a microarray analysis, we found that hsa_circ_0003315—a circRNA located at chr16:74493579–74497377, 477 nucleotides in length, and formed by back-splicing from exons 5–8 of the Golgi glycoprotein 1 (GLG1) gene—was upregulated in CRC tissues compared to that in adjacent normal tissues (data not shown); therefore, we have named this circRNA, circ_GLG1. To date, no reports have described a functional role or associated mechanism for circ_GLG1 in CRC. In this study, we investigated the roles of circ_GLG1 in CRC to determine its functional mechanism. Our study will provide a basis for further elucidating the mechanism of CRC occurrence and development, as well as that of a new diagnostic biomarker and potential therapeutic target for patients with CRC.
Materials and Methods
CRC Tissue Samples
Forty pairs of CRC tissues and matched non-tumor tissues were obtained from patients at The Second Affiliated Hospital of Jilin University (Changchun, China) between December 2017 and March 2019. None of the patients underwent chemotherapy or radiotherapy before surgery. Both cancer and normal tissues were immediately snap-frozen and stored at −80°C in an ultra-low temperature freezer until further analysis. The study was approved by the Ethics Committee of The Second Affiliated Hospital of Jilin University (approval number 2020–077). Informed consent forms were signed by all individuals prior to acquiring and studying their tissues.
Cell Culture
Human CRC cell lines (HCT116, SW620, and DLD1 cells) and the human normal colon epithelial cell line (NCM460) were purchased from the Shanghai Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences, Shanghai, China). Cells were cultured in RPMI 1640 medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin solution (Gibco). All cell lines were maintained at 37°C in a 5% CO2 atmosphere. Exponentially growing cells were used for the experiments.
RNA Extraction and Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA was extracted from clinical tissues and cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s protocol. Total RNA was quantified using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Total RNA purity was evaluated based on the ratio of the optical density (OD) at 260 nm to that at 280 nm. Total RNA (1 µg) was converted to complementary DNA using a First Strand cDNA Synthesis kit (TransGen Biotech, Beijing, China), and quantitative PCR (qPCR) was performed using TransStart® Top Green qPCR SuperMix (TransGen Biotech, Beijing, China) on an Applied Biosystems 7500 Sequence Detection System (Thermo Scientific). Relative expression levels were calculated using the 2−ΔΔCt method.21 The mRNA expression levels of β-actin were used to normalize the relative expression levels of circRNAs and mRNAs and the levels of U6 were used to normalize miRNA expression levels. The primers used for qRT-PCR analysis are shown in Table 1.
 |
Table 1 Primers Used for Quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR) Analysis |
Cell Transfections
Two small-interfering RNAs (siRNAs) against circ_GLG1 (siRNA-1 and siRNA-2), a circ_GLG1 negative-control siRNA (siRNA control), and an miR-622 inhibitor were purchased from RiboBio (Shanghai, China). The sequences are shown in Table 2. For the transfections, DLD1 cells were seeded into 6-well plates at a density of 5 × 105 cells/well and cultured for 24 h, after which they were transfected with siRNA(s) and/or the miR-622 inhibitor using Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions. The final concentrations of the siRNAs and miR-622 inhibitor used for transfections were 40 nM and 100 nM, respectively.
 |
Table 2 The Sequences of the siRNA or miRNA Inhibitor |
Cell Counting Kit-8 (CCK-8) Assay
Transfected DLD1 cells were plated in 96-well plates at a density of 2.0 × 103 cells/well. The cells were cultured for 24, 48, 72, or 96 h before 10 μL of CCK-8 solution (Dojindo, Kumamoto, Japan) was added to the culture medium in each well. After incubation for an additional 2 h at 37°C, the OD at 450 nm was measured using a multifunctional microplate reader (Bio-Rad Laboratories, Hercules, CA, USA). The experiment was repeated three times.
Colony-Formation Assay
The proliferation rates of transfected DLD1 cells were determined using a colony-formation assay. Briefly, transfected DLD1 cells were seeded into 35-mm cell culture dishes at a density of 2000 cells/well and cultured for 2 weeks at 37°C with 5% CO2. Then, the colonies were fixed with 4% paraformaldehyde for 15 min and stained with 1% crystal violet for 10 min at room temperature. The colonies were counted and photographed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The experiment was repeated three times.
Transwell Assay
The cell-invasion ability of transfected DLD1 cells was determined using 24-well transwell chambers pre-coated with Matrigel (Corning, Corning, NY, USA). Briefly, at 24 h post-transfection, DLD1 cells (2 × 104) in 200 μL serum-free medium were seeded into the upper compartment and 700 μL culture medium containing 10% FBS was added to the lower compartment. After 24 h, the cells located on the upper surfaces of the transwell chamber were removed with cotton swabs and the cells located on the lower surfaces were fixed at room temperature for 15 min with 4% paraformaldehyde and then stained for 15 min with 0.5% crystal violet. Five random areas of the membrane were photographed and used for cell counting using an inverted microscope (Olympus, Tokyo, Japan). The experiment was repeated three times.
Wound-Healing Assay
Cell-migration abilities were assessed by performing a wound-healing assay. Transfected DLD1 cells were cultured in 6-well plates until 100% confluency was reached, after which a straight scratch with a width of approximately 0.8 mm was made in the center of each well using a sterile 1-mL pipette tip. After the cells were scratched (0 h), detached cells were washed away with phosphate-buffered saline (PBS), and the remaining cells were cultured at 37°C for 24 h. Images were taken at 0 and 24 h, using microscopy. For each image, the distance between both sides of the scratch was measured using ImageJ software (National Institutes of Health). The experiment was repeated three times.
Western Blot Analysis
When transfected DLD1 cells, cultured in 6-well plates, reached 80% confluence, the medium was aspirated and cells were washed with pre-chilled 1× PBS. Then, the DLD1 cells in each well were lysed with 200 μL RIPA buffer (Beyotime Biotechnology, Shanghai, China) and 2 μL phenylmethyl sulfonic acid (Beyotime Biotechnology). The protein concentrations in the cell lysates were measured using a BCA Protein Assay kit (Beyotime Biotechnology), according to the manufacturer’s protocol. Proteins were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Millipore, Bedford, MA, USA). After blocking the membranes in low-fat milk for 2 h, they were incubated with a primary antibody against Kirsten rat sarcoma (KRAS) (1:1000 dilution; Affinity Biosciences, OH, USA) or β-actin (1:2000 dilution; Abcam, Cambridge, UK) overnight at 4°C and at 25°C for 2 h with a horseradish peroxidase-conjugated secondary antibody (1:5000 dilution, ZSGB-BIO, Beijing, China). The membranes were washed and visualized using enhanced chemiluminescence (Thermo Scientific) and the bands were quantified using ImageJ software (National Institutes of Health). β-Actin protein expression was used as an internal control. The experiment was repeated three times.
Dual-Luciferase Reporter Assay
Based on the complementary region of circ_GLG1/KRAS and miR-622, wild-type dual-luciferase reporter plasmids (pEZX-MT06-Wt-circ_GLG1 and pEZX-MT06-Wt-KRAS) and mutant plasmids (pEZX-MT06-Mut-circ_GLG1 and pEZX-MT06-Mut-KRAS) were synthesized by RiboBio. The sequences of the four plasmids are shown in Table S1. For the luciferase reporter assays, DLD1 cells were seeded into 24-well plates and cultured for 24 h. The reporter plasmid and miR-622 or negative-control (NC) mimic were co-transfected into DLD1 cells using Lipofectamine 2000 (Invitrogen). The sequences for miR-622 mimic were 5′-ACA GUC UGC UGA GGU UGG AGC-3′ (sense) and 5′-UCC AAC CUC AGC AGA CUG UAA-3′ (antisense). The sequences for NC mimic were 5′-UAU UGC ACU CUC CCC GGC CUG A-3′ (sense) and 5′-ACA GGC CGG GGG AGA GUG AAU A-3′ (antisense). After incubation for 48 h, luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA), according to the manufacturer’s instructions. Renilla luciferase activities were normalized to Firefly luciferase activities.
Statistical Analysis
Statistical analysis of the results was performed using IBM SPSS software, Version 22 (IBM Corp., Armonk, NY, USA) and the results are presented as the means ± standard deviation. Student’s t-test was used to calculate P values when comparing two independent groups. Associations between clinicopathological features and circ_GLG1 expression were evaluated by performing chi-square tests. P < 0.05 was considered to indicate a statistically significant difference.
Results
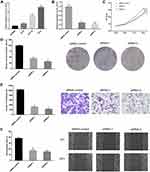
Circ_GLG1 Was Upregulated in CRC Tissues
Circ_GLG1 was formed via back-splicing from exons 5–8 of the GLG1 gene, with a length of 477 nucleotides. The structure of circ_GLG1 is shown in Figure 1A. We detected circ_GLG1 expression in 40 pairs of CRC tissues and adjacent normal tissues. The results showed that the expression of circ_GLG1 was significantly upregulated (P < 0.05) in the CRC tissues (Figure 1B), and 75% (30/40) of the CRC patients showed increased expression of circ_GLG1 in tumor tissues compared with that in the adjacent normal tissues (Figure 1C). Circ_GLG1 expression was not associated with clinical-pathological characteristics, including age, sex, lactation status, or tumor parameters (differentiation, invasion, stage, and lymph node metastasis) in patients with CRC (Table S2).
Circ_GLG1 Was Upregulated in CRC Cells and circ_GLG1 Silencing Inhibited CRC Cell Proliferation, Invasion, and Migration
We detected circ_GLG1 expression levels in three colon cancer cell lines (HCT8, HCT116, and DLD1 cells) and a normal human colonic epithelial cell line (NCM460 cells). The results showed that circ_GLG1 expression levels were higher in all three colon cancer cell lines than in NCM460 cells (Figure 2A). The highest circ_GLG1 expression level was found in the DLD1 cell line; therefore, we used the DLD1 cell line for subsequent experiments. Two circ_GLG1-specific siRNAs (siRNA-1 and siRNA-2) and a control siRNA were transfected into the DLD cells. The qRT-PCR results showed that, compared with the siRNA control, both siRNA-1 and siRNA-2 significantly downregulated (P < 0.05) circ_GLG1 expression in DLD1 cells (Figure 2B).
In the CCK-8 experiments, we found that DLD1 cells transfected with circ_GLG1 siRNAs had significantly lower proliferation rates (P < 0.05) than cells transfected with the control siRNA (Figure 2C). Similarly, DLD1 cell clone formation was significantly inhibited (P < 0.05) in the circ_GLG1 siRNA transfectants compared to that in the control-siRNA transfectant (Figure 2D). Further, we performed transwell and wound-healing assays to analyze the effect of circ_GLG1 on cell invasion and migration. The results showed that downregulating circ_GLG1 significantly decreased (P < 0.05) the invasion and migration abilities of DLD1 cells (Figure 2E and F).
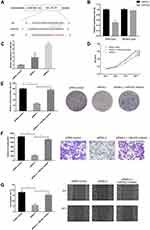
Circ_GLG1 Promoted CRC Cell Proliferation and Invasion by Regulating miR-622 Expression
Considering that the major function of circRNAs in various cancers is serving as miRNA sponges, we predicted that some miRNAs could potentially bind to circ_GLG1. The online software program, Circular RNA Interactome,22 predicted that the potential targets of circ_GLG1 comprise a total of 12 miRNAs including miR-622, with the selection criteria of context score percentile ≥ 90, as shown in Table S3. A luciferase vector encoding circ_GLG1 was constructed with or without the miR-622-binding site (referred to here as the wild-type and mutant vector, respectively). The results of dual-luciferase reporter assay showed that, after normalization to the control group, the luciferase activity decreased significantly (P < 0.05) in cells co-transfected with an miR-622 mimic and the wild-type circ_GLG1 vector, but not the mutant circ_GLG1 vector (Figure 3A and B). Furthermore, downregulation of circ_GLG1 expression with siRNA-1 or siRNA-2 led to upregulation of miR-622 expression (P < 0.05, Figure 3C).
To further confirm that circ_GLG1 plays a biological role by regulating miR-622 expression, we co-transfected DLD1 cells with an miR-622 inhibitor and circ_GLG1 siRNA. As seen from the results of the CCK-8 assay, the proliferation ability of DLD1 cells significantly decreased (P < 0.05) after transfection with siRNA-2, but not after co-transfecting siRNA-2 and the miR-622 inhibitor (P < 0.05, Figure 3D). Similar results were obtained by performing colony-formation assays (P < 0.05, Figure 3E). In the transwell assays, transfection of siRNA-2 alone reduced the number of cells that crossed the membrane. However, after co-transfection with the miR-622 inhibitor, the number of cells that passed though the membrane significantly increased (P < 0.05) to a level comparable to that of the control group, thus eliminating the negative effects of siRNA-2 (Figure 3F). Similar results were obtained in wound-healing assays, which were used to detect cell-migration abilities (P < 0.05, Figure 3G).
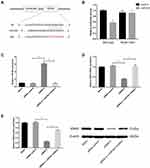
Circ_GLG1 Regulates CRC via the circ_GLG1–miR-622–KRAS Axis
As circRNA–miRNA–mRNA axes have been found to play important roles in various cancers, we used the online software program, miRWalk2.0,23 to identify a predicted binding site of miR-622 in the KRAS gene transcript. The dual-luciferase reporter assay showed that the luciferase activity significantly decreased in cells co-transfected with miR-622 mimics and a wild-type KRAS vector, but not a mutant KRAS vector (P < 0.05, Figure 4A and B).
To validate the circRNA–miRNA–mRNA axis in DLD1 cells, we knocked down circ_GLG1 expression, which was subsequently transfected with an miR-622 inhibitor. The qRT-PCR results showed that miR-622 expression was significantly lower (P < 0.05) in cells co-transfected with the miR-622 inhibitor and siRNA-2 than in cells transfected with siRNA-alone (Figure 4C). Then, we detected KRAS expression by performing qRT-PCR and Western blotting assays. The results showed that the relative expression of KRAS at both the mRNA and protein levels significantly decreased in circ_GLG1-silenced cells, and this effect was abolished in the presence of an miR-622 inhibitor (P < 0.05, Figure 4D and E).
Discussion
CRC seriously endangers human health.1 Therefore, the identification of new targets and molecular mechanisms is critical for the early detection and treatment of CRC. Here, we studied a circRNA termed circ_GLG1 and systematically elucidated its role in CRC. We found that circ_GLG1 was significantly overexpressed in CRC tissues and three CRC cell lines. In in vitro experiments, we showed that circ_GLG1 knockdown inhibited DLD1 cell proliferation, invasion, and migration. Mechanistically, we found that circ_GLG1 promoted the proliferation and invasion of CRC cells through the circ_GLG1–miR-622–KRAS axis.
We first measured circ_GLG1 expression levels in 40 pairs of CRC tissues and matched non-tumor tissues. The results showed that circ_GLG1 expression was upregulated in CRC tissues compared to that in matched non-tumor tissues, indicating that circ_GLG1 could be used as a potential diagnostic biomarker. To verify its function, we detected circ_GLG1 expression in three colon cancer cell lines (HCT116, HCT8, and DLD1 cells) and one normal human intestinal epithelial cell line (NCM460 cells). The results showed that circ_GLG1 expression was higher in all three colon cancer cell lines than in the normal intestinal epithelial cell line. Subsequently, we found that silencing circ_GLG1 expression inhibited the growth and invasion of CRC cells by performing CCK-8, colony-formation, transwell, and wound-healing assays. The above experiments confirmed the carcinogenic effect of circ_GLG1 in CRC.
The known mechanisms of circRNAs include the following: (1) CircRNAs can act as miRNA sponges;24 (2) CircRNAs can regulate the transcription of host genes by interacting with RNA polymerase II and cis-acting elements;25 (3) Some circRNAs can bind to RNA-binding proteins and regulate their functions;26 (4) In addition to acting as non-coding RNAs, some circRNAs have a conserved open reading frame that can be translated into proteins that play functional roles;27 Among the abovementioned biological functions, the sponge function of miRNAs is the most important discovered to date. Therefore, we questioned whether circ_GLG1 exerts its oncogenic effects by sponging one or more miRNAs.
The regulation between circRNAs and miRNAs is complex. A circRNA molecule can combine with multiple miRNAs. In addition, a single miRNA may be regulated via multiple circRNAs at the same time.28 Because the interaction between circRNA and miRNA occurs through complementary hybridization, databases have been developed that can predict miRNA targets based on their sequences.29 In this study, using the Circular RNA Interactome online tool to predict the potential miRNA-binding sites in circ_GLG1, we obtained a total of 12 miRNAs. Among them, miR-622 attracted our attention, mainly because it is closely related to the occurrence and development of various cancers.30–34 It has been reported that miR-622 expression is significantly downregulated in melanoma cells and tissues, which correlates with the overall survival of patients with melanoma.30 In addition, miR-622 has been identified as a tumor suppressor gene in patients with hepatocellular carcinoma,31 gastric cancer,32 colorectal cancer,33 and glioma.34 In this study, we first confirmed the existence of an miR-622-binding site in circ_GLG1 by performing dual-luciferase reporter assays. Then, we showed that miR-622 expression increased after DLD1 cells were transfected with circ_GLG1 siRNA. Furthermore, the miR-622 inhibitor reversed the inhibition of proliferation, invasion, and migration caused by the low expression of circ_GLG1, suggesting that circ_GLG1 exerted its biological functions via miR-622 in CRC.
Through the online software miRWalk2.0, we found that KRAS is one of the potential binding targets of miR-622. Accumulating evidence have shown that miR-622 can inhibit cell proliferation, migration, and invasion by targeting KRAS in melanoma,35 glioblastoma,36 and CRC.37 KRAS is a member of the RAS oncogene family.38 As downstream signaling molecules of the epidermal growth factor receptor, RAS proteins are involved in many key cellular processes, including cell-cycle regulation, cell proliferation, cell survival, regulation of inflammatory responses, and vesicular transport.39 In CRC, KRAS mutations drive tumorigenesis and are associated with the efficacy of targeted therapy.40 It has been confirmed that KRAS expression can be regulated via some miRNAs in different tumors. For example, miR-316 can inhibit the proliferation and invasion of malignant melanoma cells by inhibiting KRAS expression and the activity of the AKT–ERK signaling pathway.41 MiR-532-5p promotes apoptosis of lung adenocarcinoma cells by targeting KRAS mRNA.42 Overexpressing miR-19a inhibits KRAS expression and angiogenesis in CRC cells, which in turn inhibits tumor proliferation and invasion.43 In this study, we first confirmed the existence of an miR-622-binding site in KRAS mRNA by performing dual-luciferase reporter assays. Then, we showed that downregulating circ_GLG1 expression caused KRAS downregulation at both the mRNA and protein levels and that this effect was abolished in the presence of an miR-622 inhibitor, suggesting that circ_GLG1 exerted its biological function via the miR-622–KRAS axis in CRC.
In conclusion, the results of this study confirmed that circ_GLG1 was upregulated in CRC tissues and serves as an oncogene to promote the proliferation and invasion of CRC cells. Mechanistically, we confirmed that circ_GLG1 promoted the proliferation and invasion of CRC cells by regulating the miR-622–KRAS axis. This study provides useful information for further elucidating the molecular mechanism of CRC tumorigenesis and may provide leads to develop new diagnostic biomarkers and potential therapeutic targets for patients with CRC. However, some aspects of this study were not very rigorously assessed. Although we confirmed that circ_GLG1 could bind to miR-622, the mechanism of circRNAs is complex and it is not clear whether circ_GLG1 could also bind to other miRNAs or some RNA binding proteins. Furthermore, in the future, we plan to verify the regulation of circ_GLG1 on tumors in vivo, using mouse tumor models.
Abbreviations
circRNA, circular RNA; circ_GLG1, circRNA derived from golgi glycoprotein 1 mRNA; CRC, colorectal cancer; KRAS, Kirsten rat sarcoma; miRNA, microRNA; GLG1, golgi glycoprotein 1; qRT-PCR, quantitative reverse transcriptase-polymerase chain reaction; siRNAs, small-interfering RNAs; CCK-8, cell counting kit-8; PBS, phosphate-buffered saline.
Acknowledgments
This work was supported by the Education Department of Jilin Province, P.R.C. [grant number JJKH20190059KJ], the Science and Technology Department of Jilin Province, P.R.C. [grant number 20190201050JC], and the Science and Technology Department of Jilin Province, P.R.C. [grant number 20200201543JC].
Disclosure
The authors declare no competing financial and/or non-financial interests in relation to the work described.
References
1. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi:10.3322/caac.21395
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
3. Chen WQ, Zheng RS, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
4. Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging. 2016;11:967–976. doi:10.2147/CIA.S109285
5. Greene J, Baird AM, Brady L, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi:10.3389/fmolb.2017.00038
6. Yao R, Zou H, Liao W. Prospect of Circular RNA in hepatocellular carcinoma: a novel potential biomarker and therapeutic target. Front Oncol. 2018;8:332.
7. Wang M, Yu F, Circular LP. RNAs: characteristics, function and clinical significance in hepatocellular carcinoma. Cancers (Basel). 2018;10:8. doi:10.3390/cancers10080258
8. Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11(1):98.
9. Su C, Han Y, Zhang H, et al. CiRS-7 targeting miR-7 modulates the progression of non-small cell lung cancer in a manner dependent on NF-κB signalling. J Cell Mol Med. 2018;22(6):3097–3107. doi:10.1111/jcmm.13587
10. Ji F, Du R, Chen T, et al. Circular RNA circSLC26A4 accelerates cervical cancer progression via miR-1287-5p/HOXA7 axis. Mol Ther Nucleic Acids. 2019;19:413–420. doi:10.1016/j.omtn.2019.11.032
11. Fu Y, Cai L, Lei X, et al. Circular RNA ABCB10 promotes hepatocellular carcinoma progression by increasing HMG20A expression by sponging miR-670-3p. Cancer Cell Int. 2019;19:338. doi:10.1186/s12935-019-1055-z
12. Pan Z, Lin J, Wu D, et al. Hsa_circ_0006948 enhances cancer progression and epithelial-mesenchymal transition through the miR-490-3p/HMGA2 axis in esophageal squamous cell carcinoma. Aging (Albany NY). 2019;11(24):11937–11954. doi:10.18632/aging.102519
13. Xu ZQ, Yang MG, Liu HJ, et al. Circular RNA hsa_circ_0003221 (circPTK2) promotes the proliferation and migration of bladder cancer cells. J Cell Biochem. 2018;119(4):3317–3325. doi:10.1002/jcb.26492
14. Pan H, Li T, Jiang Y, et al. Overexpression of circular RNA ciRS-7 abrogates the tumor suppressive effect of miR-7 on Gastric Cancer via PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 2018;119(1):440–446. doi:10.1002/jcb.26201
15. Xu T, Wu J, Han P, et al. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680. doi:10.1186/s12864-017-4029-3
16. Wang Y, Mo Y, Gong Z, et al. Circular RNAs in human cancer. Mol Cancer. 2017;16(1):25. doi:10.1186/s12943-017-0598-7
17. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi:10.1016/j.cell.2009.01.002
18. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035
19. Han J, Zhao G, Ma X, et al. CircRNA circ-BANP-mediated miR-503/LARP1 signaling contributes to lung cancer progression. Biochem Biophys Res Commun. 2018;503(4):2429–2435. doi:10.1016/j.bbrc.2018.06.172
20. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305–317. doi:10.1016/j.canlet.2017.06.027
21. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
22. Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13(1):34–42.
23. Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi:10.1038/nmeth.3485
24. Yang L, Fu J, Zhou Y. Circular RNAs and their emerging roles in immune regulation. Front Immunol. 2018;9:2977. doi:10.3389/fimmu.2018.02977
25. Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264. doi:10.1038/nsmb.2959
26. Yang Q, Du WW, Wu N, et al. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24(9):1609–1620. doi:10.1038/cdd.2017.86
27. Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. doi:10.1038/cr.2017.31
28. Yamamura S, Imai-Sumida M, Tanaka Y, Dahiya R. Interaction and cross-talk between non-coding RNAs. Cell Mol Life Sci. 2018;75:467–484. doi:10.1007/s00018-017-2626-6
29. Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42.
30. Dietrich P, Kuphal S, Spruss T, et al. MicroRNA-622 is a novel mediator of tumorigenicity in melanoma by targeting Kirsten rat sarcoma. Pigment Cell Melanoma Res. 2018;31(5):614–629.
31. Song WH, Feng XJ, Gong SJ, et al. microRNA-622 acts as a tumor suppressor in hepatocellular carcinoma. Cancer Bio Ther. 2015;16(12):1754–1763.
32. Guo XB, Jing CQ, Li LP, et al. Down-regulation of miR-622 in gastric cancer promotes cellular invasion and tumor metastasis by targeting ING1 gene. World J Gastroenterol. 2011;17(14):1895–1902. doi:10.3748/wjg.v17.i14.1895
33. Fang Y, Sun B, Li Z, et al. MiR-622 inhibited colorectal cancer occurrence and metastasis by suppressing K-Ras. Mol Carcinogen. 2016;55(9):1369–1377. doi:10.1002/mc.22380
34. Zhang R, Luo H, Wang S, et al. MiR-622 suppresses proliferation, invasion and migration by directly targeting activating transcription factor 2 in glioma cells. J Neuro-Oncol. 2015;121(1):63–72. doi:10.1007/s11060-014-1607-y
35. Dietrich P, Kuphal S, Spruss T, Hellerbrand C, Bosserhoff AK. MicroRNA-622 is a novel mediator of tumorigenicity in melanoma by targeting Kirsten rat sarcoma. Pigment Cell Melanoma Res. 2018;31(5):614–629.
36. Han Z, Yang Q, Liu B, et al. MicroRNA-622 functions as a tumor suppressor by targeting K-Ras and enhancing the anticarcinogenic effect of resveratrol. Carcinogenesis. 2012;33(1):131–139. doi:10.1093/carcin/bgr226
37. Sun Y, Li Z, Chen Z, Xiang J. Mir-622 inhibited colorectal cancer occurrence and metastasis by suppressing K-Ras. Mol Carcinog. 2016;55(9):1369–1377.
38. Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004(250):RE13.
39. Collins MA, Bednar F, Zhang Y, et al. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J Clin Invest. 2012;122(2):639–653. doi:10.1172/JCI59227
40. Allegra CJ, Rumble RB, Hamilton SR, et al. Extended ras gene mutation testing in metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy: american Society of Clinical Oncology Provisional Clinical Opinion Update 2015. J Clin Oncol. 2016;34:179–185. doi:10.1200/JCO.2015.63.9674
41. Kang K, Zhang J, Zhang X, et al. MicroRNA-326 inhibits melanoma progression by targeting KRAS and suppressing the AKT and ERK signalling pathways. Oncol Rep. 2018;39(1):401–410.
42. Griesing S, Kajino T, Tai MC, et al. Thyroid transcription factor-1-regulated microRNA-532-5p targets KRAS and MKL2 oncogenes and induces apoptosis in lung adenocarcinoma cells. Cancer Sci. 2017;108(7):1394–1404. doi:10.1111/cas.13271
43. Chen M, Lin M, Wang X. Overexpression of miR-19a inhibits colorectal cancer angiogenesis by suppressing KRAS expression. Oncol Rep. 2018;39(2):619–626.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.