Back to Journals » Infection and Drug Resistance » Volume 15
A Case of Meningitis in an Infant Due to Hypervirulent Klebsiella pneumoniae Transmission Within a Family
Authors Zhang Z , Wen H, Wang H, Zhang P, Li J, Liang Y, Liu Y, Sun L, Xie S
Received 2 June 2022
Accepted for publication 24 August 2022
Published 29 August 2022 Volume 2022:15 Pages 4927—4933
DOI https://doi.org/10.2147/IDR.S376055
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Zongwei Zhang, Hainan Wen, Hui Wang, Pan Zhang, Jing Li, Yueyi Liang, Yanchao Liu, Lihong Sun, Shoujun Xie
Department of Clinical Laboratory, The Affiliated Hospital of Chengde Medical University, Chengde, Hebei, 067000, People’s Republic of China
Correspondence: Shoujun Xie, Department of Clinical Laboratory, The Affiliated Hospital of Chengde Medical University, Chengde, Hebei, 067000, People’s Republic of China, Tel +86 15633142883, Email [email protected]
Abstract: Hypervirulent Klebsiella pneumoniae (hvKP), an emerging pathotype derived from K. pneumoniae, frequently causes invasive infections of multiple organs and is associated with both high disability and fatality rates. In this study, a case of meningitis in a young infant caused by hvKP is presented. Cytological and biochemical examinations of the cerebrospinal fluid (CSF) revealed purulent meningitis, a diagnosis that was confirmed by a positive CSF culture result. The pathogen was identified as hvKP through analysis of positive virulence-associated genes. Meanwhile, hvKP was also isolated from stool samples of both the infant and her father. Antimicrobial susceptibility, capsular typing, and multilocus sequence typing (MLST) of three isolates from the infant’s CSF and stool and her father’s stool samples were analyzed. The three K. pneumoniae isolates were susceptible to all antibiotics except ampicillin and were identified as capsular serotype K2 and sequence type 86. These genetic relatedness analyses indicated that the strain isolated from the infant’s CSF might have originated from her father’s stool via familial transmission. This case is the first report of meningitis in an infant due to hvKP transmitted within a family.
Keywords: hvKP, purulent meningitis, infant, familial transmission
Introduction
Klebsiella pneumoniae is an opportunistic pathogen that causes a wide range of infections (including pneumonia) in addition to bloodstream and urinary tract infections. Hypervirulent K. pneumoniae is an invasive variant that differs from the classical K. pneumoniae (cKP) due to its hypermucoviscosity and hypervirulence.1 Over the past three decades, hvKP has emerged as a clinically important pathogen that can cause highly invasive infections, such as pyogenic liver abscess, endophthalmitis, meningitis, and necrotizing fasciitis, in healthy and immunocompromised individuals.2 Bacterial meningitis is a major cause of morbidity and mortality in young children.3 In Chinese infants less than three months of age, the main pathogens that cause meningitis are Escherichia coli, Enterococcus faecium, and Staphylococcus epidermidis.4 Documentation of KP meningitis in children is sparse with only a few case reports concerning children under the age of 1 year.5–7 A report of meningitis caused by hvKP in infants has not been reported yet. Recently, a young infant with meningitis seemingly caused by hvKP via familial transmission presented to our hospital. Familial spread was speculated after analyzing the genetic relatedness of the isolates from the patient’s and her family members’ specimens. To the best of our knowledge, this report describes the first case of meningitis in an infant due to hvKP spread within a family. By reporting such a rare case, we aim to attract the attention of professionals in hospitals and other health-care establishments in addition to social communities to the potential transmission mechanism of hvKP and take the necessary prevention and control steps to prevent further spread of hvKP strains.
Case Presentation
On February 16, 2022, a one-month-old Chinese infant girl was admitted to our hospital with a 16.5 h history of fever and cough. On admission, the patient had a fever of 38.1 ℃ without chills and a tic, accompanied by poor mental response and irritability. Her anterior fontanelle was bulging resulting from an increase in pressure in the brain cavity, and a cranial slit separation could be felt. The breath sounds from both lungs were thick, and sounds associated with phlegm could be heard. Bilateral Babinsiki signs were positive, and Brudzinski signs were negative. Laboratory testing revealed elevated levels of C-reactive protein (18.23 mg/L; reference:< 6 mg/L), procalcitonin (0.12 ng/mL; reference:< 0.05 ng/mL) and interleukin-6 (153.90 pg/mL; reference: < 7 pg/mL). Her white blood cell count ([WBC] 10.90 x 109/L) was within the reference range, but the ratio of neutrophils to white blood cells (63.4%) showed a slight elevation. Result from brain computed tomography (CT) was unremarkable, while a lung CT scan revealed inflammation. On the evening of admission, she underwent a lumbar puncture, and CSF was obtained for cytological and biochemical examinations and microbial cultures. Blood, urine, and sputum cultures were also obtained at the same time. The WBC count of the CSF showed a remarkable increase (8800 x 106/L; segmented neutrophils, 90%), accompanied by elevated CSF protein level (> 3.00 g/L; reference: 0.2–0.45 g/L) and decreased CSF glucose level (< 1.11 mmol/L; reference: 1.9–4.7 mmol/L). Analysis of the CSF confirmed the diagnosis of purulent meningitis after which empirical antibiotic therapy with intravenous meropenem and vancomycin was initiated. On the third day of admission, the CSF culture result was found to be positive, and the pathogen was preliminarily identified as hypermucoviscous K. pneumoniae based on a positive string test (Figure 1). Antimicrobial susceptibility testing suggested that the isolate was susceptible to all antibiotics with the exception of natural resistance to ampicillin (Table 1). Blood, urine, and sputum cultures exhibited negative results after five days of culture. After 20 days of anti-infection treatment with meropenem, the next two consecutive CSF cultures were negative, the patient’s condition improved, and she was discharged from the hospital on March 9.
 |
Table 1 Susceptibility Profiles of Klebsiella Pneumoniae Isolated from the Infant’s CSF, Stool Samples, and the Father’s Stool Sample |
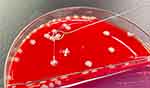 |
Figure 1 The string test in an isolated colony of Klebsiella pneumoniae from the CSF on an agar plate, which showed hypermucoviscosity with a > 5-mm-long viscous filament. |
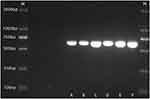
To trace the origin of the central nervous system infection in the infant, her clinician was questioned, and stool samples from the infant and her parents were cultured. K. pneumoniae was isolated from the stool samples of both the infant and her father, and the string test of these two isolates were both positive. The antimicrobial susceptibility of three isolates from the infant’s CSF and stool samples and her father’s stool were characterized using the VITEK-2 system. Polymerase chain reaction (PCR) was used to detect five virulence-associated genes, including rmpA, iroB, peg-344, iucA, and rmpA2, from the three isolates. Genetic relatedness was analyzed using capsular typing by PCR and multilocus sequence typing. Antimicrobial susceptibility testing suggested that the three isolates were susceptible to all antibiotics except ampicillin (Table 1). Virulence gene results show that the isolates from the CSF and stool samples from the infant were positive for peg-344, iroB, and rmpA and negative for rmpA2 and iucA, whereas the isolate from the father’s stool was positive for all the five virulence genes (Figure 2). The three K. pneumoniae isolates were all identified as capsular serotype K2 (Figure 3) and ST86.
Materials and Methods
Bacterial Isolates and Identification
The CSF sample was centrifuged at 3000 rpm for 10 min, the supernatant was discarded, the precipitates were inoculated on Blood Agar (BIO-KONT, Wenzhou, China), China Blue Lactose Agar (BIO-KONT, Wenzhou, China) and Chocolate Agar (BIO-KONT, Wenzhou, China), and incubated at 37 ℃ for 24 h. The stool samples were directly inoculated on Blood Agar, China Blue Lactose Agar, and Salmonella Shigella agar (BIO-KONT, Wenzhou, China) at 37 ℃ for 24 h. Species identification was confirmed by matrix-assisted laser desorption-ionization time-of-flight mass spectrometry (MALDI-TOF-MS, BioMérieux, France), and the identification of each strain was repeated in triplicate.
Antibiotic Susceptibility Testing
The antibiotic susceptibility of K. pneumoniae isolates were determined using the Vitek 2 Compact system (BioMérieux, France) according to Clinical Laboratory Standards Institute guidelines.8 Details of the antibiotics used for this test are listed in Table 1.
String Test
Single colonies cultured on blood agar plates were obtained and tested for their capabilities to form viscous strings. Colonies were touched with a loop, and the loop was then lifted vertically from the surface of the agar plate.9 A positive result was defined by the formation of viscous strings > 5 mm in length.
DNA Extraction
DNA extraction was performed using the boiling method. Some colonies from the surface of blood agar were suspended in 200 µL of sterile water, prepared for 0.5 McFarland bacterial suspension, and boiled for 10 min at 100 ℃. The bacterial suspension after boiling was centrifuged at 13,000 rpm for 5 min, total DNA was extracted from the supernatant, and then stored at −20 ℃ for amplification.
Capsular Typing and Virulence-Associated Genes
PCR (DNA Engine Peltier Thermal Cycle Meter, MJ Research, United States) was used to detect capsular types and virulence-associated genes, including rmpA, iroB, peg-344, iucA, and rmpA2. Primer sequences and reaction conditions were referenced from relevant literature.10,11 The total PCR reaction system was 50 uL, including 2 uL upstream and downstream primers, 2 uL DNA template, 19 µL sterile ddH2O, 25 µL 2×Mix. The amplification products were separated on a 2% agarose gel (Bioweste Regular Agarose G-10, Spain), and a single band indicated the corresponding gene is positive.
Multilocus Sequence Typing
MLST was carried out for three K. pneumoniae isolates. Amplification of the seven housekeeping genes, rpoB, gapA, mdh, pgi, PhoE, infB, and tonB was done using the primer pairs as described earlier.12 The total PCR reaction system was performed as described above. Amplification was performed using DNA Engine Peltier Thermal Cycle Meter (MJ Research, United States) applying a specific program: (1) DNA pre-degeneration at 94 ℃, 5 min; (2) DNA denaturation at 94 ℃, 1 min; (3) DNA renaturation at 60 ℃, 1 min; (4) extension at 72 ℃, 1 min; (5) 30 cycles at 94 ℃, 1 min; 60 ℃, 1 min; 72 ℃, 1 min; (6) final extension at 72 ℃, 10 min; and (7) reaction termination at 4 ℃. The amplicons were separated in 1.5% agarose gel (Bioweste Regular Agarose G-10, Spain) and sequenced by Sanger sequencing (ABI 3730XL DNA Analyzer, United States). The resulting sequences were submitted to the K. pneumoniae MLST database (https://pubmlst.org/bigsdb?db=pubmlst_mlst_seqdef) and obtained the STs of each isolate.
Discussion
Hypervirulent K. pneumoniae is an emerging and evolving pathotype. In recent years, an increasing trend in severe infections caused by hvKP has appeared. The environment possibly acts as a reservoir for human infection by K. pneumoniae, either through colonization or infection.13 Among body parts, gastrointestinal colonization is likely a common and crucial reservoir in terms of risk of transmission and infection.14 In general, as with opportunistic K. pneumoniae, hvKP infections can also be caused by endogenous colonizing strains.13 Several potential sources of transmission of K. pneumoniae have been identified. Person-to-person contact is one of the most common modes of transmission,15 and contaminated surfaces and apparatus have been identified as other routes of dissemination.16 The possibility of fecal–oral transmission has also been proposed on the basis of molecular typing of isolates from family members and the environment as described in a study from Taiwan.17 In our case, isolates from the infant’s CSF and stool samples and her father’s stool sample were all identified as hvKP, and the antibiotic susceptibility, capsular types, and multilocus sequence typing exhibited the same characteristics. It seems that the K. pneumoniae isolate that had colonized in the father’s gut was transmitted to the infant by means of household contact after which they colonized in the infant’s gut. The strain entered the bloodstream and crossed the blood–brain barrier while the infant was in a state of compromised immunity after which it infected the central nervous system. The familial spread of hvKP isolates causing liver abscesses was first reported in Japan in 2011,18 and osteomyelitis caused by hvKP spread within a family has also been previously reported.19 Our report is the first case of purulent meningitis due to hvKP dissemination within the same household.
At present, no international consensus for the identification of hvKP exists. The hypervirulence of hvKp strains is mediated by genes on a large virulence plasmid20 or within chromosomal islands.21 Detection of virulence-related genes by PCR is the most commonly used method to identify hvKP. The five virulence genes detected in our study were found in a pLVPK virulence plasmid, which harbors a series of virulence-associated genes, including the capsular polysaccharide synthesis regulator rmpA, its homolog rmpA2, and multiple iron acquisition systems, including iucABCDiutA and iroBCDN siderophore gene clusters,22 and peg-344, a putative metabolite transporter.23 Russo et al10 used epidemiological and experimental lines of evidence to identify several laboratory markers for identifying hvKp strains and found that peg-344, iroB, iucA, rmpA, and rmpA2 all identified an isolate as a member of the hvKp strain cohort with an accuracy of >0.95. Virulence plasmids undergo constant genetic changes during the transmission process to adapt to different host environments.24 Some plasmids have been found to contain gene deletions during the process of transmission, presumably due to evolution of the strain during culture in the laboratory25 or some form of selective pressure, such as antibiotic-related or environmental.26 It is possible that some genes that do not play a role in virulence expression and plasmid propagation may have been lost during transmission.27 The role of the virulence plasmid is strain-dependent and associated with the context of K. pneumoniae isolates and the plasmid determinants of hypervirulence retain the potential for microevolution.26 In our report, the strain isolated from the infant’s specimens were positive for three of the five genes in a plasmid, while the strain isolated from her father’s sample was positive for all five virulence genes. The reason for this phenomenon is likely due to loss of the region that carried the other two genes during transmission of the plasmid due to some form of selective pressure, such as an antibiotic-related or environmental.
HvKP is mainly prevalent among Asians, especially the Chinese. In a study concerning K. pneumoniae colonization of the intestinal tract of healthy Chinese adults, the rate of K. pneumoniae isolation from stool specimens was up to 62.1% and of serotype K1/K2, which are thought to be the major virulence determinants, comprising 9.8% of all the K. pneumoniae strains.28 Chinese ethnicity itself might be a major element predisposing an individual to intestinal colonization by hvKP strains.28 The vast majority of K1 isolates belong to the sequence type 23 (ST23), a finding that highly correlates with a suppurative liver abscess.29 K2 is more widespread than K1 for causing community-acquired extrahepatic abscess.30 Lin et al reported that ST65 and ST86 are the most common types in K2 isolates and these two major MLST groups were also the most prevalent STs of K2 in stool carriage isolates from carriers without a history of KP liver abscess.31 The hvKP isolates from CSF and stools in our report were identified as K2-ST86, which may partly further confirm the perspective that K2 is more inclined to cause extrahepatic infection and capable of colonizing in the gut under normal conditions.
Conclusion
To the best of our knowledge, this case is the first report of familial spread of K. pneumoniae K2-ST86 isolate causing purulent meningitis. Based on the high prevalence rate of hvKP in the Chinese population and the potential spreading mechanisms via close contact and fecal–oral transmission, attention to this issue should be raised among public health-care professionals and social communities. In addition, necessary measures should be taken to prevent the hvKP strains from further disseminating through the community and hospital settings.
Ethics and Consent
This report has been approved by the Affiliated Hospital of Chengde Medical University’s ethics committee to publish the case details. Written informed consent to have the case details published has been provided by the patient’s mother.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Liu Z, Gu Y, Li X, et al. Identification and characterization of NDM-1-producing Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae in China. Ann Lab Med. 2019;39(2):167–175. doi:10.3343/alm.2019.39.2.167
2. Hu D, Li Y, Ren P, et al. Molecular epidemiology of hypervirulent carbapenemase-producing Klebsiella pneumoniae. Front Cell Infect Microbiol. 2021;11:661218. doi:10.3389/fcimb.2021.661218
3. Bingen E, Levy C, de la Rocque F, et al. Bacterial meningitis in children: a French prospective study. Clin Infect Dis. 2005;41(7):1059–1063. doi:10.1086/432944
4. Peng X, Zhu Q, Liu J, et al. Prevalence and antimicrobial resistance patterns of bacteria isolated from cerebrospinal fluid among children with bacterial meningitis in China from 2016 to 2018: a multicenter retrospective study. Antimicrob Resist Infect Control. 2021;10(1):24. doi:10.1186/s13756-021-00895-x
5. Sundaram V, Agrawal S, Chacham S, Mukhopadhyay K, Dutta S, Kumar P. Klebsiella pneumoniae brain abscess in neonates: a report of 2 cases. J Child Neurol. 2010;25(3):379–382. doi:10.1177/0883073809338326
6. Biswas B, Mondal M, Thapa R, Mallick D, Datta AK. Neonatal brain abscess due to extended-spectrum beta-lactamase producing Klebsiella pneumoniae. J Clin Diagnostic Res. 2014;8(11):Pd01–02. doi:10.7860/JCDR/2014/10160.5198
7. Khaneja M, Naprawa J, Kumar A, Piecuch S. Successful treatment of late-onset infection due to resistant Klebsiella pneumoniae in an extremely low birth weight infant using ciprofloxacin. J Perinatol. 1999;19(4):311–314. doi:10.1038/sj.jp.7200084
8. CLSI M100-Ed31. Performance Standards for Antimicrobial Susceptibility Testing. Washington DC: Clinical and Laboratory Standards Institute; 2021.
9. Yu VL, Hansen DS, Ko WC, et al. Virulence characteristics of Klebsiella and clinical manifestations of K. pneumoniae bloodstream infections. Emerg Infect Dis. 2007;13(7):986–993. doi:10.3201/eid1307.070187
10. Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9). doi:10.1128/JCM.00776-18
11. Turton JF, Perry C, Elgohari S, Hampton CV. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59(Pt 5):541–547. doi:10.1099/jmm.0.015198-0
12. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J Clin Microbiol. 2005;43(8):4178–4182. doi:10.1128/JCM.43.8.4178-4182.2005
13. Martin RM, Bachman MA. Colonization, Infection, and the Accessory Genome of Klebsiella pneumoniae. Front Cell Infect Microbiol. 2018;8:4. doi:10.3389/fcimb.2018.00004
14. Dorman MJ, Short FL. Genome watch: Klebsiella pneumoniae: when a colonizer turns bad. Nat Rev Microbiol. 2017;15(7):384. doi:10.1038/nrmicro.2017.64
15. Casewell M, Phillips I. Hands as route of transmission for Klebsiella species. Br Med J. 1977;2(6098):1315–1317. doi:10.1136/bmj.2.6098.1315
16. Jarvis WR, Munn VP, Highsmith AK, Culver DH, Hughes JM. The epidemiology of nosocomial infections caused by Klebsiella pneumoniae. Infect Control. 1985;6(2):68–74. doi:10.1017/S0195941700062639
17. Chiu CH, Su LH, Wu TL, Hung IJ. Liver abscess caused by Klebsiella pneumoniae in siblings. J Clin Microbiol. 2001;39(6):2351–2353. doi:10.1128/JCM.39.6.2351-2353.2001
18. Harada S, Tateda K, Mitsui H, et al. Familial spread of a virulent clone of Klebsiella pneumoniae causing primary liver abscess. J Clin Microbiol. 2011;49(6):2354–2356. doi:10.1128/JCM.00034-11
19. Lee CH, Chae JD, Choe W, et al. Osteomyelitis caused by hypervirulent Klebsiella pneumoniae: the first Korean case with family spread. Ann Lab Med. 2021;41(2):250–254. doi:10.3343/alm.2021.41.2.250
20. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/S1473-3099(17)30489-9
21. Lin TL, Lee CZ, Hsieh PF, Tsai SF, Wang JT. Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J Bacteriol. 2008;190(2):515–526. doi:10.1128/JB.01219-07
22. Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi:10.1016/j.gene.2004.05.008
23. Bulger J, MacDonald U, Olson R, Beanan J, Russo TA. Metabolite transporter PEG344 is required for full virulence of hypervirulent Klebsiella pneumoniae strain hvKP1 after pulmonary but not subcutaneous challenge. Infect Immun. 2017;85(10). doi:10.1128/IAI.00093-17
24. Lev AI, Astashkin EI, Kislichkina AA, et al. Comparative analysis of Klebsiella pneumoniae strains isolated in 2012–2016 that differ by antibiotic resistance genes and virulence genes profiles. Pathog Glob Health. 2018;112(3):142–151. doi:10.1080/20477724.2018.1460949
25. Lery LM, Frangeul L, Tomas A, et al. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol. 2014;12:41. doi:10.1186/1741-7007-12-41
26. Li P, Liang Q, Liu W, et al. Convergence of carbapenem resistance and hypervirulence in a highly-transmissible ST11 clone of K. pneumoniae: an epidemiological, genomic and functional study. Virulence. 2021;12(1):377–388. doi:10.1080/21505594.2020.1867468
27. Yang X, Dong N, Chan EW, Zhang R, Chen S. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2021;29(1):65–83. doi:10.1016/j.tim.2020.04.012
28. Lin YT, Siu LK, Lin JC, et al. Seroepidemiology of Klebsiella pneumoniae colonizing the intestinal tract of healthy Chinese and overseas Chinese adults in Asian countries. BMC Microbiol. 2012;12:13. doi:10.1186/1471-2180-12-13
29. Liao CH, Huang YT, Chang CY, Hsu HS, Hsueh PR. Capsular serotypes and multilocus sequence types of bacteremic Klebsiella pneumoniae isolates associated with different types of infections. Eur J Clin Microbiol Infect Dis. 2014;33(3):365–369. doi:10.1007/s10096-013-1964-z
30. Ku YH, Chuang YC, Yu WL. Clinical spectrum and molecular characteristics of Klebsiella pneumoniae causing community-acquired extrahepatic abscess. J Microbiol Immunol Infect. 2008;41(4):311–317.
31. Lin JC, Koh TH, Lee N, et al. Genotypes and virulence in serotype K2 Klebsiella pneumoniae from liver abscess and non-infectious carriers in Hong Kong, Singapore and Taiwan. Gut Pathog. 2014;6:21. doi:10.1186/1757-4749-6-21
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.