Back to Journals » Cancer Management and Research » Volume 11
5-Aza-CdR Regulates RASSF1A By Inhibiting DNMT1 To Affect Colon Cancer Cell Proliferation, Migration And Apoptosis
Authors Chen J, Wu L, Xu H, Cheng S
Received 3 September 2019
Accepted for publication 22 October 2019
Published 8 November 2019 Volume 2019:11 Pages 9517—9528
DOI https://doi.org/10.2147/CMAR.S229726
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Antonella D'Anneo
Jinyuan Chen,1 Lixiang Wu,2 Hong Xu,3 Shubang Cheng1
1Department of Gastrointestinal Surgery, People’s Hospital of Longhua, Shenzhen, Guangdong 518109, People’s Republic of China; 2Guangzhou Concord Cancer Center, Guangzhou, Guangdong 510620, People’s Republic of China; 3Department of Pharmacy, People’s Hospital of Longhua, Shenzhen, Guangdong 518109, People’s Republic of China
Correspondence: Shubang Cheng
Department of Gastrointestinal Surgery, People’s Hospital of Longhua, Shenzhen, Guangdong 518109, People’s Republic of China
Email [email protected]
Objective: To evaluate 5-Aza-CdR’s inhibited effects on migration, proliferation, and apoptosis in colon cancer cells and its potential mechanisms.
Methods: HCT-116, SW480, and SW620 were divided into HCT116 group, HCT116+5-Aza-CdR group, SW480 group, SW480+5-Aza-CdR group, SW620 group and SW620+5-Aza according to experimental needs. MTT test was chosen to investigate cell proliferation; Transwell test was used to evaluate cell migration; scratch assay was used to investigate cell invasion; flow cytometry was used to investigate apoptosis; immunofluorescence assay was used to investigate the protein level of DNMT1 and RASSF1A in cells; qRT-PCR was used to examine DNMT1, RASSF1A, RAS, Raf1, MEK, Grb2 and ERK transcription levels.
Results: Compared with HCT116 group, 5-Aza-CdR+HCT116 group inhibited cell proliferation, increased apoptosis rate, decreased invasive ability, decreased DNMT1 expression, increased expression of RASSF1A, decreased expression of RAS, Raf1, MEK, Grb2 and ERK. SW480 was compared with 5-Aza-CdR+SW480 group and SW620 group with 5-Aza-CdR+SW620 group. Their change trend of detection index was similar to that in HCT-116 group and HCT116+5-Aza-CdR group.
Conclusion: 5-Aza-CdR can obviously inhibit the proliferation, migration and invasion of three colon cancer cell lines. Its mechanism maybe relies on the inhibition of DNMT1 mRNA level and protein level and the enhancement of RASSF1A mRNA level and protein level.
Keywords: DNA methylation, colon cancer, RASSF1A, 5-Aza-CdR, DNMT1
Introduction
Colon cancer is a highly aggressive malignancy that accounts for about 10% of cancer-triggered mortality.1 Approximately 3–5% of colon cancers worldwide are caused by genetic factor, and up to 25% of patients have family susceptibility, but most colon cancers are sporadic without sensitive genes.2 More and more studies have proved that epigenetic changes, such as DNA methylation, hydromethylation, chromatin remodeling, histone methylation and acetylation RNA interference, also play a critical part in the occurrence and development of colon cancer, expect as genetically unstable phenotypes such as chromosome and microsatellite instability.3,4
Epigenetic changes are characterized by dynamic changes compared with genetic defects and can be affected by age, environment, lifestyle and dietary habit. Abnormal DNA methylation includes focal hypermethylation of the promoter CpG island and global hypomethylation of the DNA. The former will trigger transcriptional silencing of the target gene, while the latter will reduce the stability of the chromosome, which is the most critical epigenetic experiment in the development of cancer.5,6 It has been reported that RASSF1A, an important tumor suppressor gene, is present in a highly methylated form in colon cancer. Maintenance of DNA methylation status is primarily dependent on DNA methyltransferase function (DNMT).
DNMT1 is a major DNMTs involved in the maintenance of methylation in DNA replication. Moreover, the status of DNA methylation is also closely related to the extracellular signal-regulated kinase (ERK) pathway. ERK activation can trigger further excitation of downstream Ras protein, and the activated Ras-ERK signaling pathway is a key event in regulating cell proliferation and apoptosis.7,8 Studies have shown that up-regulation of DNMT1 can inhibit the expression of RASSF1A,9–11 while over-expression of RASSF1A can inhibit cell proliferation, enhance cell migration and apoptosis12–14
Given the potentially reversible nature of DNA methylation changes, which often precede genetic events in multistage colon cancer, this presents an anticipated and hopeful opportunity for cancer prevention and treatment. At present, there are many studies exploring the treatment of cancer by inhibiting the abnormal methylation of DNA in cancer cells. 5-Azacytidine and 5-aza-29-deoxycytidine (5-Aza-CdR) have been determined to be valid DNMT inhibitors. 5-Aza-CdR can directly bind to DNA single strands during DNA replication, and on the other hand, it can catalyze proteasome-dependent degradation of demethylated DNMT1, and finally exert demethylation biological activity.15–17 At present, 5-Aza-CdR has been widely used in the therapy of hematological tumors, but its application in colon cancer is still rare, and its mechanism is still unclear.
In our study, the inhibitory function of 5-Aza-CdR on proliferation and migration of three colon cancer cell lines was firstly investigated, and its mechanism was closely related to the inhibition of DNMT1 and the increase of RASSF1A transcription level. The above results can provide new ideas and strategies for the colon cancer’s clinical therapy.
Materials And Methods
Materials And Reagent
Human colon cancer cells HCT-116, SW480, SW620 were purchased from the Shanghai Cell Bank of the Chinese Academy of Sciences; 5-Azacytidine (100 mg; Ab142744) was purchased from Abcam; The cell culture reagent was prepared from the following reagents: DMEM-high glucose medium (Hyclone, Cat. No. SH30022.01B), Streptomycin (Hyclone, Cat. No. SH30010), PBS potassium phosphate buffer (Hyclone, Cat. No. SH30256.01B), fetal bovine serum (Hyclone, Cat. No. SH30087.01).
Experimental Grouping
According to the experimental needs, the three cell lines, HCT116, SW480 and SW620, were divided into HCT116 group, HCT116+5-Aza-CdR group, SW480 group, SW480+5-Aza-CdR group, SW620 group and SW620+5-Aza-CdR group. The water bath was preheated to 37°C, and the super-clean workbench that was irradiated with ultraviolet light for 30 mins was wiped with 75% alcohol. Disinfected centrifuge tubes, straws, culture flasks, etc., were placed in order on the workbench. Remove the cell culture flask and perform aseptically. Aspirate the old culture solution. The trypsin-EDTA solution (1mL/25cm2) was added and slightly wash the bottom. Aspirated the trypsin-EDTA solution and placed it in a 37°C incubator for 2–3 mins. Gently pated the wall of the flask to allow most of the cells to fall off and observed under an inverted microscope. When the cells were to be separated and presented in round granules, trypsin was stopped by adding an appropriate amount of fresh serum-containing medium. The pipette was sucked up and down several times to break up the cell mass. After mixing evenly, made up 3n (n is the number of passage bottles) mL medium (MEM), and transferred to a new culture bottle according to the dilution ratio. And then put in CO2 incubator (5% CO2, saturated humidity, 37°C). The concentration of 5-Aza-CdR was 5 μmol/L and the treatment time was 24 hrs.
MTT Assay For Cell Proliferation
The cells of each treatment group were taken for the following experiment. After digesting the cells, the cells were blown, counted to modulate the cell concentration to 1×105 cells/mL, and then divided into 96-well plates, 100 µL per well, namely, 1×104 cells per well. The adherent cells were collected at various time points for detection after cell adhesion Cells were harvested at different time points (0, 24, 48, 72 hrs) and added to cell Titer 96 AQ One Solution Cell Proliferation assay reagent (Promega, Cat. No. G3582) at a ratio of 1/10. That was, 10 μL of the detection solution was added to 100 μL of the culture solution. After 4 hrs of incubation, the plate was read by an enzyme-labelling measuring instrument and the OD490 data were read by MTT assay. Each assay was performed in triplicate.18
Flow Cytometer Detection Of Apoptosis
The medium from each of the cell culture plates was transferred to a 15 mL tube. Cells in the plate were gently swashed with PBS. 0.5 mL 0.25% trypsin without EDTA to incubate until they began to fall off from the culture plate wall under the microscope. Afterwards, the cells were completely exfoliated from the culture plate wall by gentle continuous beating. The cells were suspended gently in step 1 medium or pre-cooled 1x binding buffer to make their density approximately 1x 106 cells/mL. Transferred 0.5 mL of the cell suspension from the cell culture plated (5 x 105 cells) to a clean centrifuge tube. Added 1.25 μL Annexin V-FITC. 15 mins for reaction (18–24°C). The supernatant was removed in centrifuge at 1000 g for 5 mins at room temperature. The cells were slightly suspended with 0.5 mL of precooked binding buffer. Added 10 μL of Propidium Iodide. The sample was placed on ice to preserve it from light, and analyzed by flow cytometry at once.19
qRT-PCR Detection
According to the manufacturer’s protocol, total RNA using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was separated. By using reverse transcription kit (Takara, Dalian, China) isolated RNA reverse transcribed into cDNA. The results were normalized according to the expression of glyceraldehyde 3-phosphate dehydrogenase (GAPDH). PCR primers are shown in Table 1. qRT-PCR data collection device on the ABI 7500 (Applied Biosystems, Foster City, CA, USA) was carried out. PCR reaction was carried out at 50°C 2 mins, at 95°C 10 mins, at 95°C 15 s, at 60°C 30 s. A relative expression level of the gene using 2−△△Ct method was calculated. All the PCR-reaction was repeated in a qRT triplicate experiments.
 |
Table 1 The Forward And Reverse Primers Used In Real-Time RT-PCR |
Cellular Immunofluorescence
The cells in the culture medium were fixed in 4% paraformaldehyde for 30 mins; washed 3 times with PBS for 5 mins each time; 0.2% Triton X-100 for 5 mins; washed 3 times with PBS for 5 mins each time; 10% normal goat serum was blocked for 0.5 hrs. Then, the primary antibody was incubated overnight at 4°C (anti-RASSF1a antibody [3F3], 1:500; 1:500; ab23950; anti-Dnmt1 antibody [EPR3521(2)], 1:250, ab134148); washed 3 times with PBS for 5 mins each time. Fluorescently labeled secondary antibody (1:200) was incubated for 60 mins in the dark at room temperature; washed 3 times with PBS for 5 mins each time. DAPI was incubated for 5 mins in the dark; washed once with PBS for 5 mins and washed twice for 5 mins each time; sealed with fluorescent anti-quenching agent; microscopic examination was done and photos were taken.20
Data Processing And Statistical Analysis
Student's T-test was carried out with SPSS 21.0 software, and GraphPad prism6.0 was used for drawing. All data were presented as mean ± standard error. When P < 0.05, the difference was considered statistically significant.
Results
Effect Of 5-Aza-CdR On Proliferation Of Colon Cancer Cells
MTT assayed the effect of 5-Aza-CdR on the proliferation of three colon cancer cells, HCT116, SW480 and SW620. As the experimental results are shown in Figure 1, compared with the HCT116 group, the cells proliferation of the HCT116 group treated with 5-Aza-CdR was inhibited (Figure 1A). In addition, compared with SW480 group, the cells proliferation of SW480 treated with 5-Aza-CdR group was inhibited (Figure 1B). Moreover, compared with the SW620 group, the cells proliferation of SW620 group treated with 5-Aza-CdR was inhibited (Figure 1C). Above experimental results showed that 5-Aza-CdR could obviously suppress the proliferation of colon cancer cells.
Effect Of 5-Aza-CdR On Invasion Of Colon Cancer Cells
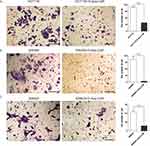
Transwell experiments were used to test the influence of 5-Aza-CdR on the invasion of three colon cancer cells, HCT116, SW480 and SW620. The experimental results are shown in Figure 2. The invasive ability of 5-Aza-CdR dealing with HCT116 cells was decreased compared with the HCT116 group (Figure 2A). In addition, compared with the SW480 group, the invasive ability of 5-Aza-CdR dealing with SW480 group cells was decreased (Figure 2B). Moreover, the invasive ability of 5-Aza-CdR dealing with SW620 cells was decreased compared with the SW620 group (Figure 2C). The above experimental results suggested that 5-Aza-CdR significantly inhibited the invasion of colon cancer cells.
Effect Of 5-Aza-CdR On Migration Of Colon Cancer Cells
The influence of 5-Aza-CdR on the migration of three colon cancer cells, HCT116, SW480 and SW620, was investigated by scratch test. The experimental results are shown in Figure 3. Compared with the HCT116 group, the mobility of 5-Aza-CdR dealing with HCT116 cells was significantly decreased (Figure 3A). In addition, compared with the SW480 group, the mobility of the 5-Aza-CdR dealing with SW480 group was significantly decreased (Figure 3B). Moreover, compared with the SW620 group, the mobility of 5-Aza-CdR dealing with SW620 cells was significantly decreased (Figure 3C). The above experimental results indicated that 5-Aza-CdR obviously inhibited the migration of colon cancer cells.
Effect Of 5-Aza-CdR On Apoptosis Of Colon Cancer Cells
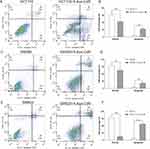
Flow cytometry was chosen to investigate the influence of 5-Aza-CdR on apoptosis of HCT116, SW480 and SW620 colon cancer cells. The experimental results are shown in Figure 4. The apoptosis rate of the 5-Aza-CdR dealing with HCT116 group was enhanced compared with the HCT116 group (Figure 4A and B). In addition, compared with the SW480 group, the apoptosis rate of the 5-Aza-CdR dealing with SW480 group was increased (Figure 4C and D). Moreover, the apoptosis rate of the 5-Aza-CdR dealing with SW620 group was enhanced compared with the SW620 group (Figure 4E and F). The above results suggested that 5-Aza-CdR triggered apoptosis in three colon cancer cells, but the specific mechanism remained unclear.
Effect Of 5-Aza-CdR On The Expression Of DNMT1 And RASSF1A Proteins In Colon Cancer Cells
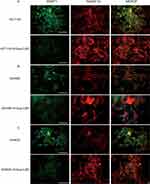
Immunofluorescence experiments were performed to evaluate the effects of 5-Aza-CdR on the protein level of DNMT1 and RASSF1A in three colon cancer cells, HCT116, SW480 and SW620. The results of the immunization experiment are shown in Figure 5. Compared with the HCT116 group, the protein level of DNMT1 was decreased and the protein level of RASSF1A was increased in HCT116 group with the treatment of 5-Aza-CdR (Figure 5A); compared with SW480 group, the protein level of DNMT1 was decreased and the protein level of RASSF1A was increased in SW480 group treated with 5-Aza-CdR (Figure 5B); compared with SW620 group, protein level of DNMT1 was decreased and protein level of RASSF1A was increased in SW620 group treated with 5-Aza-CdR (Figure 5C). The above experimental results indicated that 5-Aza-CdR restored abnormally elevated DNMT1 levels and abnormally reduced RASSF1A levels in colon cancer cells.
Effects Of 5-Aza-CdR On The Transcriptional Levels Of DNMT1, RASSF1A, ERK, MEK, Grb2, Raf1 And RAS In Colon Cancer Cells
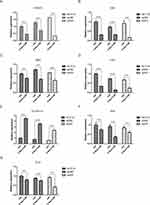
In order to further explore the mechanism of 5-Aza-CdR in colon cancer cells' apoptosis, the mRNA level of DNMT1, RASSF1A, ERK, MEK, Grb2, Raf1 and RAS was tested by qRT-PCR. All results are shown in Figure 6. Compared with HCT116 group, the mRNA level of DNMT1 was decreased, the mRNA level of RASSF1A was elevated, and the mRNA level of RAS, Raf1, MEK, Grb2 and ERK was decreased in the HCT116 group treated with 5-Aza-CdR. In addition, compared with the SW480 group, the mRNA level of DNMT1 was decreased, the mRNA level of RASSF1A was increased, and the mRNA level of RAS, Raf1, MEK, Grb2 and ERK was decreased in the SW480 group treated with 5-Aza-CdR; compared with SW620 group, the mRNA level of DNMT1 was decreased, the mRNA level of RASSF1A was increased, and the mRNA level of RAS, Raf1, MEK, Grb2 and ERK was decreased in the SW620 group treated with 5-Aza-CdR. Together, these results suggested that 5-Aza-CdR could increase the biological efficacy of colon cancer cell apoptosis by inhibiting the ERK-Ras signaling pathway.
Discussion
In this research, the benefit effects of 5-Aza-CdR on three colon cancer cell proliferation and apoptosis were systematically investigated, and its mechanism may be closely related to inhibition of DNA methylation and ERK-Ras signaling pathway. Based on the results of this study, we have provided new and powerful evidence for the clinical use of 5-Aza-CdR for colon cancer treatment.
In recent years, many studies have pointed out that the development of cancer is closely related to genetic factors and epigenetic modifications. Epigenetic modification refers to a heritable phenotypic change that occurs when the gene does not change in sequence. DNA methylation is an important event in epigenetic modification. The DNA methylation is mainly catalyzed by DNMT and methylated by S-adenosylmethionine (SAM). This change is often reversible. Currently known DNMTs include DNMT1, DNMT3a, and DNMT3b. DNMT1 is mainly involved in newly synthesized single-stranded DNA methylation and transmits methylation information to progeny cells. DNMT3a and DNMT3b are mainly responsible for DNA methylation during embryogenesis.21 The high expression of DNMTs in cancer cells leads to hypermethylation of the tumor-suppressor gene and then resulting in inactivation, ultimately leading to tumor formation. Numerous studies have found that DNMT1 is associated with abnormal DNA methylation, both of which are closely related to the occurrence and development of tumors.22 Dong et al also have found that cancer can be treated by inhibiting DNMT1.23 This is because the abnormal expression of DNMT1 in the G0/G1 phase of cell cycle can silence the tumor suppressor and eliminate the normal blocking signal, leading to the uncontrolled growth of cancer cells.24 At the same time, inhibiting DNMT1 activity can reduce the methylation of inhibiting genes, promote their re-expression, and reverse the phenotype of malignant tumors.25
5-Aza-CdR is the most widely used demethylated nucleoside analogue and plays a major part in the demethylation process by suppressing the expression and activation of low concentrations of DNMTs.26 Masumeh et al have reported that 5-Aza-CdR can up-regulate the expression of p21, p27 and p57 genes, thereby enhancing the apoptosis and cell growth inhibition of colon cancer cells.14 Liu et al have found that 5-Aza-CdR can inhibit the growth and proliferation of HXO-RB44 retinoblastoma cells.27 Nikbakht et al have found that 5-Aza-CdR can induce death and apoptosis of human pancreatic cancer cell lines by activating RASSF1A and up-regulating Bax gene.28 And, 5-Aza-CdR has been studied in oral squamous cell carcinoma, bladder cancer, pancreatic cancer, lung adenocarcinoma, endometrial cancer.29–33 In this study, we tested the mRNA levels of DNMT1 in three colon cancer cells by immunofluorescence and qRT-PCR. The results indicated that DNMT1 was highly expressed in the three colon cancer cells. But the colon cancer cell DNMT1 treated with 5-Aza-CdR showed a significant decrease in translational level and transcriptional level. In addition, 5-Aza-CdR is also used in different cancers.
RASSF1A is an important tumor-suppressor genes in cells. RASSF1A-mediated protein expression mainly works on the cell signal transduction pathway associated with Ras protein, inducing cell apoptosis and blocking the synthesis of endogenous cyclin which affects proliferation. In the study of metastatic colon cancer, Romano et al have found that RASSF1A protein is at low expression level in all stages of colon cancer development, and the related proteins Ras, Raf1, ERK and MEK in Ras signaling pathway are highly expressed at the transcriptional level.34 In the meantime, RASSF1A protein level was also markedly decreased after VEGF stimulated normal cells.35 In addition, Lorenzatoet al have found significantly lower expression of RASSF1A in other colon cancer tissues. In this experiment, we observed by immunofluorescence and qRT-PCR that compared with the protein level of colon cancer cell RASSF1A, the colon cancer cell RASSF1A treated with 5-Aza-CdR indicated an obvious increase in transcriptional level and translational level. Similarly, the mRNA levels of ERK-Ras signaling pathway-related proteins Ras, Raf1, ERK and MEK in colon cancer cells treated with 5-Aza-CdR were significantly decreased. MTT results found that 5-Aza-CdR markedly inhibited the proliferation of colon cancer cells, and flow cytometry experiments also demonstrated that 5-Aza-CdR significantly promoted apoptosis in colon cancer cells. In addition, both Transwell and scratch experiments indicated that 5-Aza-CdR obviously reduced the ability of invasion and migration of colon cancer cells.
In conclusion, 5-Aza-CdR can increase the protein and mRNA level of PASSF1A by suppressing the expression of DNMT1, and finally, trigger apoptosis of colon cancer cells by affecting the ERK-Ras signaling pathway. These results provide new insights into the clinical therapy of colon cancer, and understanding the epigenetic demethylation pathway is an important mechanism of action.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Lamm DL. Cancer statistics. CA Cancer J Clin. 2010;40(5):318–319. doi:10.3322/canjclin.40.5.318
2. Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21–33. doi:10.1038/nrg1748
3. Feinberg AP. Epigenetics at the epicenter of modern medicine. JAMA. 2008;299(11):1345. doi:10.1001/jama.299.11.1345
4. Wong JJL, Hawkins NJ, Ward RL. Colorectal cancer: a model for epigenetic tumorigenesis. Gut. 2007;56(1):140–148. doi:10.1136/gut.2005.088799
5. Peto J. Cancer epidemiology in the last century and the next decade. Nature. 2001;411(6835):390–395. doi:10.1038/35077256
6. Suzuki K, Suzuki I, Leodolter A, et al. Global dna demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9(3):199–207. doi:10.1016/j.ccr.2006.02.016
7. Goel A, Nagasaka T, Arnold CN, et al. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132(1):127–138. doi:10.1053/j.gastro.2006.09.018
8. Rodriguez J, Frigola J, Vendrell E, et al. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66(17):8462–9468. doi:10.1158/0008-5472.CAN-06-0293
9. Bai J, Zhang X, Hu K, et al. Silencing DNA methyltransferase 1 (DNMT1) inhibits proliferation, metastasis and invasion in ESCC by suppressing methylation of RASSF1A and DAPK. Oncotarget. 2016;7(28):44129–44141. doi:10.18632/oncotarget.v7i28
10. Qiu X, Zhang L, Lu S, et al. Upregulation of DNMT1 mediated by HBx suppresses RASSF1A expression independent of DNA methylation. Oncol Rep. 2014;31(1):202–208. doi:10.3892/or.2013.2848
11. Agarwal S, Amin KS, Jagadeesh S, et al. Mahanine restores RASSF1A expression by down-regulating DNMT1 and DNMT3B in prostate cancer cells. Mol Cancer. 2013;12(1):99. doi:10.1186/1476-4598-12-99
12. Bao Y, Liu X, Liu Y, et al. Ras-association domain family 1 (RASSF1A) gene regulates progression, migration and invasion of bladder cancer. Surg Oncol. 2019;30:63–71. doi:10.1016/j.suronc.2019.05.009
13. Boyanapalli SS, Li W, Fuentes F, et al. Epigenetic reactivation of RASSF1A by phenethyl isothiocyanate (PEITC) and promotion of apoptosis in LNCaP cells. Pharmacol Res. 2016;114:175–184. doi:10.1016/j.phrs.2016.10.021
14. Sanaei M, Kavoosi F. Effect of 5-Aza-2ʹ-deoxycytidine in comparison to valproic acid and trichostatin A on histone deacetylase 1, DNA methyltransferase 1, and CIP/KIP family (p21, p27, and p57) genes expression, cell growth inhibition, and apoptosis induction in colon cancer SW480 cell line. Adv Biomed Res. 2019;8:52.
15. Ghoshal K, Datta J, Majumder S, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the ken box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25(11):4727–4741. doi:10.1128/MCB.25.11.4727-4741.2005
16. Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2”-deoxycytidine residues by chromatin-associated dnmt1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38(13):4313–4324. doi:10.1093/nar/gkq187
17. Schermelleh L, Spada F, Easwaran HP, et al. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat Methods. 2005;2(10):751–756. doi:10.1038/nmeth794
18. Ramiólluch L, Yeste M, Fernándeznovell JM, et al. Oligomycin a-induced inhibition of mitochondrial ATP-synthase activity suppresses boar sperm motility and in vitro capacitation achievement without modifying overall sperm energy levels. Reprod Fertil Dev. 2013;26(6):883. doi:10.1071/RD13145
19. Zhang J, Jiang Y, Zhu J, et al. Overexpression of long non-coding RNA colon cancer-associated transcript 2 is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Oncol Lett. 2017;14(6):6907–6914. doi:10.3892/ol.2017.7049
20. Tian D, Tian M, Han G, Li JL. Increased glucocorticoid receptor activity and proliferation in metastatic colon cancer. Sci Rep. 2019;9(1):1–7. doi:10.1038/s41598-019-47696-2
21. Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9(16):2395. doi:10.1093/hmg/9.16.2395
22. Kang X, Kong F, Huang K, et al. LncRNA MIR210HG promotes proliferation and invasion of non-small cell lung cancer by upregulating methylation of CACNA2D2 promoter via binding to DNMT1. Onco Targets Ther. 2019;12:3779–3790. doi:10.2147/OTT
23. Kim DH, Kim HM, Huong PTT, et al. Enhanced anticancer effects of a methylation inhibitor by inhibiting a novel DNMT1 target, CEP 131, in cervical cancer. BMB Rep. 2019;52(5):342–347. doi:10.5483/BMBRep.2019.52.5.055
24. Lee E, Wang J, Yumoto K, et al. DNMT1 regulates epithelial-mesenchymal transition and cancer stem cells, which promotes prostate cancer metastasis. Neoplasia. 2016;18(9):553–566. doi:10.1016/j.neo.2016.07.007
25. Zhang Y, Chen FQ, Sun Y-H, et al. Effects of DNMT1 silencing on malignant phenotype and methylated gene expression in cervical cancer cells. J Exp Clin Cancer Res. 2011;30:98. doi:10.1186/1756-9966-30-98
26. Baylin S, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;16(4):168–174. doi:10.1016/S0168-9525(99)01971-X
27. Liu R, Zhang XH, Zhang K, et al. 5-Aza-2-deoxycytidine inhibits retinoblastoma cell by reactivating epigenetically silenced RASSF1A gene. Int J Ophthalmol. 2014;7(1):51–56. doi:10.3980/j.issn.2222-3959.2014.01.09
28. Nikbakht DMP, Azarnezhad AP, Hashemibeni B, et al. An effective concentration of 5-Aza-CdR to induce cell death and apoptosis in human pancreatic cancer cell line through reactivating RASSF1A and up-regulation of bax genes. Iran J Med Sci. 2018;43(5):533–540.
29. do Amaral AG, Planello AC, Borgato G, et al. 5-Aza-CdR promotes partial MGMT demethylation and modifies expression of different genes in oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2019;127(5):425–432. doi:10.1016/j.oooo.2019.01.006
30. Zhang HH, Huang B, Cao YH, et al. Role of 5-Aza-CdR in mitomycin-C chemosensitivity of T24 bladder cancer cells. Oncol Lett. 2017;14(5):5652–5656. doi:10.3892/ol.2017.6853
31. Pan FP, Zhou HK, Bu HQ, et al. Emodin enhances the demethylation by 5-Aza-CdR of pancreatic cancer cell tumor-suppressor genes P16, RASSF1A and ppENK. Oncol Rep. 2016;35(4):1941–1949. doi:10.3892/or.2016.4554
32. Yang B, Yang ZG, Gao B, et al. 5-Aza-CdR can reverse gefitinib resistance caused by DAPK gene promoter methylation in lung adenocarcinoma cells. Int J Clin Exp Pathol. 2015;8(10):12961–12966.
33. Huang LP, Chen C, Wang X-P, et al. The role of 5-Aza-CdR on methylation of promoter in RASSF1A gene in endometrial carcinoma. Sichuan Da Xue Xue Bao Yi Xue Ban. 2015;46(3):380–383.
34. Romano D, Matallanas D, Weitsman G, Preisinger C, Ng T, Kolch W. Proapoptotic kinase mst2 coordinates signaling crosstalk between rassf1a, RAF-1, and akt. Cancer Res. 2010;70(3):1195–1203. doi:10.1158/0008-5472.CAN-09-3147
35. Lorenzato A, Martino C, Dani N, et al. The cellular apoptosis susceptibility cas/cse1l gene protects ovarian cancer cells from death by suppressing rassf1c. FASEB J. 2012;26(6):2446. doi:10.1096/fj.11-195982
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.