Back to Journals » OncoTargets and Therapy » Volume 12
20(R)-Ginsenoside Rg3 Influences Cancer Stem Cell Properties and the Epithelial-Mesenchymal Transition in Colorectal Cancer via the SNAIL Signaling Axis
Authors Phi LTH , Wijaya YT , Sari IN , Kim KS , Yang YG , Lee MW, Kwon HY
Received 11 June 2019
Accepted for publication 14 November 2019
Published 11 December 2019 Volume 2019:12 Pages 10885—10895
DOI https://doi.org/10.2147/OTT.S219063
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Lan Thi Hanh Phi,* Yoseph Toni Wijaya,* Ita Novita Sari, Kwang Seock Kim, Ying-Gui Yang, Min-Woo Lee, Hyog Young Kwon
Soonchunhyang Institute of Medi-Bio Science (SIMS), Soonchunhyang University, Cheonan, Republic of Korea
*These authors contributed equally to this work
Correspondence: Hyog Young Kwon; Min-Woo Lee
Soonchunhyang Institute of Medi-Bio Science (SIMS), Soonchunhyang University, 25 Bongjeong-ro, Cheonan, Chungcheongnam-do 31151, Republic of Korea
Tel +82-41-413-5021; +82-41-413-5029
Email [email protected]; [email protected]
Background: Cancer stem cells (CSCs) have been proposed as central drivers of cancer relapse in many cancers. In the present study, we investigated the inhibitory effect of 20(R)-Ginsenoside Rg3 (Rg3R), a major active component of ginseng saponin, on CSC-like cells and the Epithelial-Mesenchymal Transition (EMT) in colorectal cancer (CRC).
Methods: The effects of ginsenoside Rg3R on the colony-forming, migration, invasion, and wound-healing abilities of CRC cells were determined in HT29 and SW620 cell lines in vitro. Further, ginsenoside Rg3R was given intraperitoneally at 5mg/kg of mouse body weight to check its effect on the metastasis of CRC cells in vivo.
Results: Ginsenoside Rg3R significantly inhibited CSC properties, but did not affect cell proliferation. Moreover, ginsenoside Rg3R treatment significantly inhibited the motility of CRC cells based on migration, invasion, and wound-healing assays. The inhibitory effects of ginsenoside Rg3R on CRC are potentially mediated by significant down-regulation of the expression of stemness genes and EMT markers in CRC cells in a SNAIL-dependent manner. Furthermore, ginsenoside Rg3R treatment decreased both the number and size of tumor nodules in the liver, lung, and kidney tissues in a metastasis mouse model.
Conclusion: These findings highlighted the potential use of ginsenoside Rg3R in clinical applications for colorectal cancer treatment.
Keywords: colorectal cancer, ginsenoside Rg3R, CSCs, EMT
Introduction
Despite recent improvements in cancer treatment, colorectal cancer (CRC) remains one of the major causes of cancer-related morbidity and mortality worldwide.1,2 It has been indicated that CRC has poor prognosis when metastasized to lymph nodes or distant organs.3 Therefore, understanding the mechanisms driving metastasis is necessary for the development of new diagnostic and therapeutic strategies to improve patient outcomes in CRC. Interestingly, accumulating evidence suggested that the epithelial-mesenchymal transition (EMT) plays an important role in normal development of the colon, as well as the motility and metastasis of tumors. In addition, recent studies indicated that cancer stem cells (CSCs) are one of the central drivers of cancer relapse because of their ability to establish higher rates of proliferation, self-renewal, multi-lineage differentiation, and chemo-resistance.4–7 It has been shown that CSCs play key roles not only in the tumor progression and recurrence but also in metastasis. Activation of EMT transcription factors or induction of EMT could confer the stem-like features in cancer cells, and circulating tumor cells collected from patients with cancer metastasis have been found to express both CSC and stem cell markers.8 This relation was also indicated in colorectal cancer as colorectal CSCs have been considered as “seed” cells for invasion and metastasis in colorectal cancer.9 Therefore, there is a need to understand and target the EMT and CSCs to improve the outcomes of CRC patients.
Ginseng, a slow-growing perennial plant belonging to the genus Panax of the Araliaceae family, is one of the most well studied and commonly used traditional phytomedicines worldwide.10 Ginsenosides are the major biologically active principal components of ginseng.11 Ginsenoside Rg3, a protopanaxadiol-type ginsenoside, is a pharmacologically and physiologically active natural constituent of ginseng that has been demonstrated to exert various biological effects, including anti-cancer effect,12 and anti-inflammatory effects.13,14 Ginsenoside 20(S)-Rg3 (Rg3S) and ginsenoside 20(R)-Rg3 (Rg3R) are epimers depending on the position of the hydroxyl group (OH) on carbon-20; epimerization is produced by selective attack of the OH group after eliminating the glycosyl residue at carbon-20 during the steaming process.15 A previous study showed that ginsenoside Rg3R, but not ginsenoside Rg3S, inhibits the TGF β1-induced EMT, lung cancer migration, invasion, and anoikis resistance.16 Furthermore, ginsenoside Rg3R has been shown to inhibit cell proliferation in induced hepatocellular carcinoma in rats17 and the metastasis of B16-BL6 melanoma cells.18 However, although the stereospecific anti-tumor effects of Rg3 have been identified, the effects of ginsenoside Rg3R on CSC-like properties and EMT and the exact mechanisms underlying these effects in CRC are not fully understood.
Here, we evaluated the anti-cancer effects of ginsenoside Rg3R in CRC and the corresponding molecular mechanisms underlying these effects. Ginsenoside Rg3R was found to inhibit the CSC properties and metastasis in CRC in vitro and in vivo. The current findings not only demonstrated the anti-cancer effects of ginsenoside Rg3R, but also revealed that these effects were mediated by the downregulation of stemness markers. Further, this ginsenoside significantly inhibited the formation of tumor metastasis nodules in a mouse metastasis model. Therefore, our findings provided new insights highlighting the potential use of ginsenoside Rg3R as a novel anti-cancer agent for CRC treatment.
Materials and Methods
Reagents and Antibodies
Ginsenoside Rg3R was obtained from Ambo Institute (Korea). It was dissolved at a concentration of 20mM in DMSO as a stock solution and stored in aliquots at −20°C. Ginsenoside Rg3R was added to cell culture media to the final concentrations of 10, 50, and 100 µM for the in vitro study, and diluted with phosphate-buffered saline (PBS) to 5 mg/kg/mice for the in vivo study.
Cell Lines
The human CRC cell lines HT29 and SW620 were purchased from the Korean Cell Line Bank (KCLB). HT29 and SW620 cells were cultured in RPMI-1640 medium (Corning, USA) supplemented with 10% Fetal Bovine Serum (Corning, USA), 1% MEM essential amino acids (Corning, USA), and 1% penicillin/streptomycin (Gibco, USA) at 37°C in a humidified atmosphere containing 5% CO2. Cells were also cultured using ultra-low attachment plate in stem cell selective media including DMEM/F12 medium supplemented with 1% N2 supplement, 20 ng/mL epidermal growth factor EGF (Invitrogen, USA), and 20 ng/mL basic fibroblast growth factor (bFGF; Invitrogen, USA) and 1% penicillin/streptomycin (Gibco, USA) (Corning, USA).
Retrovirus Constructs and Transfection
Plasmids containing the open reading frame (ORF) of SNAIL (Forward, 5ʹ- AGTCCAGAATTCATGCCGCGCTCTTTCCTCGTCAGGA-3ʹ and reverse, 5ʹ- AGTCCACTCGAGTCAGCGGGGACATCCTGAGCAGCCG-3ʹ) were amplified and cloned into the MSCV-IRES-GFP vector. 293T cells were transfected with viral constructs along with Gag/pol and VSVG constructs to generate the viruses using the iN-fectTM in vitro transfection reagents (iNtRON, Korea) and following the manufacturer’s protocol. Viral supernatants were collected on days 2 and 3 after transfection and used to infect the target cells. RT-PCR and Western blotting were conducted to determine the effects of the overexpression of target genes.
RNA Extraction and Realtime PCR
RNA was isolated using Ribospin II or Hybrid R (Gene All, Korea). cDNA was converted using ReverTra Ace® qPCR Kit (TOYOBO, Japan) according to the manufacturer’s instructions. TOPreal™ qPCR 2x PreMIX (Enzynomics, Korea) were used for qPCR reaction to determine the gene expression levels of the target genes. Primer sequences for qPCR are shown in Supplementary Table 1.
Western Blotting Analysis
Cell lysates were collected and the concentration of proteins in the supernatant was determined using a Bio-Rad Protein Assay kit (Bio-Rad Laboratories, Inc., USA). Equal amounts of cell extracts (30 μg) were resolved on a 10% polyacrylamide gel and electro-transferred onto 0.45 μm hybridization nitrocellulose filter (HATF) membrane (Millipore, USA) using Trans-blot Turbo (Bio-Rad Laboratories, Inc., USA). Membranes were immunoblotted with goat polyclonal anti-Actin antibody, rabbit monoclonal anti-Snail antibody, rabbit polyclonal anti-EGFR antibody, rabbit polyclonal anti-pEGFR antibody, rabbit polyclonal anti-Akt antibody, and rabbit polyclonal anti-pAkt antibody from Cell Signaling, USA, overnight at 4°C. Membranes were incubated with either HRP-conjugated anti-rabbit immunoglobulin (Cell Signaling, USA) or HRP-linked anti-goat immunoglobulin (Santa Cruz Biotechnology, USA) for 1 hr at room temperature. Enhanced chemiluminescence (Thermo, USA) was used to detect the protein signals with the Amersham Imager 600 (GE Healthcare Life Sciences, UK).
Cell Proliferation Assay (MTT Assay)
MTT assay was performed to evaluate cell proliferation by using the Cell Proliferation Kit I according to the manufacturer’s instructions (Roche, Germany). Briefly, 5×103 cells were seeded onto a 96-well plate and incubated for 96 hrs. Cells were incubated in 5 mg/mL of MTT solution for 4 hrs and subsequently solubilized with 100 µL of solubilization solution (10% SDS in 0.01 M HCl) overnight. The absorbance was read at 575 nm and 650 nm using a plate reader.
Soft-Agar Colony-Forming Assay
Cell suspension (2×103 cells/well) and ginsenoside were mixed with 0.3% agarose in complete medium using 24-well plates coated with the 1% agarose base layer. The top of the cell layer was coated with complete medium to avoid evaporation. The number and size of colonies were determined after 15 days of incubation.
Migration and Invasion Assay
The transwell insert system (Corning, USA) with coating and without coating by 20 μL of Matrigel (BD, USA) were, respectively, used to examine the cell invasion and migration in vitro. Briefly, 100 μL of serum-free media containing 1×105 cells were seeded into each well of the insert with 600 μL of media containing 10% FBS outside of the insert. Cells were then incubated at 37°C for 18 hrs and 24 hrs in a humidified atmosphere with 5% CO2 for the migration and invasion assays, respectively. After cleaning the transwells using a cotton swap, the cells were fixed with 1% formaldehyde for 15 mins and washed with PBS 2 times. The cells were then stained with 0.1% of crystal violet for 15 mins and observed using a microscope (Leica, Germany) after washing with DW.
Wound-Healing Assay
The cells (1x105) were seeded into 24-well plate at 24 hrs before treatment. The medium was then removed when the cells reached about 90% confluence. The plates were scratched using the end of a 200 µL pipette tip (0 hr time point), and cells were washed twice with PBS to remove the loose cells. Afterwards, the cells were then treated either with or without ginsenoside Rg3R in serum-free media for 48 hrs. The images of the migrating cells were captured every 24 hrs.
Gelatin Zymography Analysis
After 96 hrs of Rg3R treatment in serum-free media, the cell supernatants were analyzed on an SDS-PAGE gel containing 0.1% gelatin. After electrophoresis, the gel was renatured twice with 2.5% Triton X-100 for 30 mins at 37°C and then washed with ddH2O. Afterwards, the gel was incubated in developing buffer (50 mM Tris-HCl pH 7.6, 50 mM NaCl, 10 mM CaCl2, 0.05% Brij 35) for 24 hrs at room temperature. The gels were then stained by Coomassie Brilliant Blue Protein Staining 1 hr and destained using destaining solution (methanol:ddH2O:acetic acid = 5:4:1) at room temperature.
Animal Experiments
HT29 CRC cells were collected and resuspended with PBS at a final concentration of 1.5×107 cells/mL. The cells (1.5×106) were intravenously injected into eight-week-old female NSG mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, The Jackson Laboratory). Mice were randomly divided into two groups (control and ginsenoside Rg3R treatment) at 48 hrs after the injection. The mice were then intraperitoneally injected with 5 mg/kg/mice ginsenoside Rg3R or an equal volume of PBS three times per week and the mice were observed daily. After 28 days, the mice were sacrificed for counting and statistical analysis of the number and weight of tumor nodules in the liver, lungs, and kidneys. Tissues were cut into 4–5 μm-thick sections, embedded in paraffin, and then stained with hematoxylin and eosin (H&E). All experimental protocols were approved by the Soonchunhyang University Institutional Animal Care and Use Committee. All animal experiments were performed in accordance with the Laboratory Animals Welfare Act.
Statistical Analysis
All experiments were independently performed at least three times. Results of RT-qPCR, Western blotting, gelatin zymography, migration, and invasion assays were analyzed using Student’s t-test. Differences were considered statistically significant at P <0.05 (*) or highly significant at P <0.01 (**).
Results
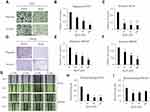
Ginsenoside Rg3R Reduces the Colony Formation Ability of CRC Cells
Ginsenoside Rg3R was isolated from Panax ginseng, and its structure was shown in Figure 1A. First, we examined the effect of ginsenoside Rg3R on the proliferation of CRC cell lines, such as HT29 and SW620. The results of MTT assay showed that ginsenoside Rg3R did not inhibit CRC cell proliferation at several concentrations (10, 50, and 100 µM ginsenoside Rg3R) after 72 hrs of incubation in liquid culture (Figure 1B and C). Next, we investigated whether ginsenoside Rg3R specifically inhibits CSC properties, even though cell proliferation of the bulk population was not affected. A recent study highlighted the emerging use of 3D cell culture, given its potential impact on early drug discovery and therapeutics for the treatment of diseases, such as cancer19 because it can more closely mimic in vivo cell environments.20 Therefore, we conducted the soft-agar colony-forming assay, which has certain advantages, including simplicity, validity, and potential for discovering new antitumor drugs,21,22 using in cancer stem cells (CSCs).23,24 Both HT29 and SW620 cell lines were treated in a layer of soft agar media containing various concentrations of ginsenoside Rg3R. Interestingly, both colony numbers and size decreased in a dose-dependent manner after 15 days of treatment with ginsenoside Rg3R (HT29, 67% at 10 µM, 81% at 50 µM, and 88% at 100 µM; SW620, 55% at 55%, 64% at 50 µM, and 81% at 100 µM) (Figure 1D and E). Therefore, the above findings suggested that ginsenoside Rg3R dramatically inhibited CSC properties in CRC.
Ginsenoside Rg3R Impairs Motility and Suppresses the EMT and CSC Signatures
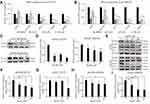
Considering that cancer cell migration and invasion are crucial for the metastatic cascade,25,26 we thus next conducted migration assay using a Matrigel-uncoated transwell system to evaluate the effects of ginsenoside Rg3R on cell motility using the CRC cell lines HT29 and SW620. The results indicated that ginsenoside Rg3R significantly attenuated the migratory ability of cancer cells by about 30% to 75% relative to the control-treated cells (HT29, 31% at 10 µM, 50% at 50 µM, and 74% at 100 µM; SW620, 43% at 10 µM, 51% at 50 µM, and 71% at 100 µM) (Figure 2A–E). In addition, we investigated the inhibitory effect of ginsenoside Rg3R on the invasive capability of CRC cells using a Matrigel-coated transwell system. Ginsenoside Rg3R significantly suppressed the invasion ability of the CRC cells by about 30% to 80% relative to control-treated cells for both CRC cell lines HT29 and SW620 after 48 hrs of treatment at several tested concentrations (HT29, 63% at 10 µM, 74% at 50 µM, and 81% at 100 µM; SW620, 27% at 10 µM, 46% at 50 µM, and 67% at 100 µM) (Figure 2A–F). Wound-healing assay was performed to further verify the inhibitory effect of ginsenoside Rg3R on the migration of CRC cells. As shown in Figure 2G–I, cells treated with ginsenoside Rg3R showed significantly lower monolayer wound-healing rates relative to cells in the control condition. The above results indicated that ginsenoside Rg3R treatment decreased the motility of CRC cells in a dose-dependent manner.
We next investigated the mechanisms by which ginsenoside Rg3R suppressed both the colony-forming ability and migratory ability of CRC cells (Figures 1 and 2). We profiled the expression signatures of genes involved in CSC stemness and the EMT; these genes have been previously established as downstream molecules involved in CSC maintenance and cancer cell mobility. To evaluate CSC properties and EMT signatures, we measured RNA and protein levels in HT29 and SW620 cells after 4 days of ginsenoside Rg3R treatment (0 to 100 µM) by RT-qPCR and immunoblots. Results showed that treatment of HT29 and SW620 cells with ginsenoside Rg3R at the indicated concentrations remarkably downregulated NANOG, OCT4, SNAIL, and MMP2 expression levels (Figure 3A and B). The protein levels of SNAIL and E-CAD, and the enzymatic activity of MMP2 were consistently and significantly reduced by the treatment with ginsenoside Rg3R (Figure 3C and Supplementary Figure 1). Therefore, the results suggested that ginsenoside Rg3R can inhibit CSC self-renewal and cancer metastasis of CRC cells by regulating the expression of CSC and EMT signatures.
Ginsenoside Rg3R Inhibits EGFR Signaling in CRC Cells
Previous studies indicated that EGFR is strongly upregulated in CRC patients, especially in stage T3 patients.27–29 Interestingly, EGFR strongly interacts with various ginsenosides, including ginsenoside Rg3; in mutant EGFR, ginsenoside Rg3 is known to interact with the GLN791 and Pro794 residues.30 Moreover, several ginsenosides, such as compound,31,32 Rb,33 Rd,34 Rh2,35 and Rg336–38 could suppress the expression level of EGFR and its signaling pathway as well. Therefore, we investigated whether ginsenoside Rg3R inhibits CRC properties by suppressing EGFR signal transduction. Thus, we examined the phosphorylation of EGFR and the downstream AKT pathway to identify the signaling pathways mediating the inhibitory effects of ginsenoside Rg3R on CRC cells. Both HT29 and SW620 cells were treated with ginsenoside Rg3R and EGF, and then the protein was extracted and analyzed by Western blotting to examine the pEGFR and pAKT levels. The pEGFR (pEGFR/tEGFR) level was reduced by about 45% and 24% relative to those in the control cells after treatment with 100 µM ginsenoside Rg3R in HT29 and SW620, respectively (Figure 3E, F, and H). Similar pattern was also observed in pAKT (pAKT/tAKT) level that was reduced by about 50% and 59% relative to those in the control cells after treatment with 100 µM ginsenoside Rg3R in HT29 and SW620, respectively (Figure 3E, G, and I). The above findings indicated that ginsenoside Rg3R suppressed the CSC-like properties and the EMT in CRC cells by inhibiting EGFR/AKT signaling.
Partial Rescue of the Inhibitory Effects of Ginsenoside Rg3R via EGF and SNAIL
To verify that ginsenoside Rg3R inhibits the migratory ability of CRC cells by downregulating EGFR/AKT signaling, we examined whether the migration ability could be restored by adding excess EGF. We evaluated the migration of HT29 cells treated with either DMSO, 50 µM of ginsenoside Rg3R alone, or 50 µM of ginsenoside Rg3R in combination with exogenous EGF. Treatment with ginsenoside Rg3R alone inhibited cancer cell migration, while combined treatment with EGF and ginsenoside Rg3R partially rescued the migratory ability of CRC cells (Figure 4A and B). In addition, downregulation of the RNA levels of the genes EGFR and SNAIL, which are downstream of EGFR signaling pathway, after ginsenoside Rg3R treatment, was dramatically abolished by EGF treatment (Figure 4C and D). This gain-of-function experiment demonstrated that the observed inhibitory effects of ginsenoside Rg3R are dependent on EGFR signaling.
Increasing evidence indicates that SNAIL plays an essential role in embryonic development and cancer progression by promoting cancer stem cell-like traits, metastasis, drug resistance, and tumor relapse.39 Our results revealed that CRC cells treated with ginsenoside Rg3R have significantly lower mRNA and protein levels of SNAIL relative to those of the control group (Figure 3). Furthermore, the inhibitory effects of ginsenoside Rg3R on SNAIL expression were partly recovered by EGF treatment. Therefore, we hypothesized that the inhibitory effects of ginsenoside Rg3R are SNAIL-dependent. To test our hypothesis, HT29 cells were transduced with either empty vector or SNAIL overexpression plasmid using a retroviral system. These cells were then treated with ginsenoside Rg3R to determine its effects on the migratory ability of cells and the expression of downstream targets. First, we confirmed SNAIL overexpression in these cells by both RT-qPCR and Western blotting (Figure 4E and F). The migratory capability of cancer cells treated with ginsenoside Rg3R was reduced to 42% of that of control cells; SNAIL overexpression rescued the inhibitory effects of ginsenoside Rg3R on cell migration, with SNAIL-overexpressing cells showing about 90% of cell migration relative to that of the control cells (Figure 4G and H). A similar pattern was also found in the colony-forming ability, with SNAIL-overexpression cells rescuing the inhibition of ginsenoside Rg3R partly (Figure 4I and J). Consistent with expected results, SNAIL overexpression also rescued the ginsenoside Rg3R-induced downregulation of downstream molecules, EGFR and FIBRONECTIN (Figure 4K). The above findings indicated that ginsenoside Rg3R also affects CSC properties and the EMT in CRC cells via SNAIL.
Ginsenoside Rg3R Reduced Metastasis of CRC Cells in vivo
The above results indicated that ginsenoside Rg3R inhibits CSC properties and the EMT in vitro. Therefore, we further examined the effects of ginsenoside Rg3R on the metastasis of CRC cells in vivo using a mouse metastasis model. After 2 days of HT29 injection into immunocompromised NSG mice, the mice were intraperitoneally injected with either ginsenoside Rg3R (5 mg/kg per mice) or PBS as control three times per week. The mice were sacrificed at 4 weeks after transplantation to evaluate tumor metastasis nodules in the liver, lungs, and kidneys (Figure 5A). The numbers of tumor metastasis nodules in the liver, lung, and kidney tissues were significantly lower in mice treated with ginsenoside Rg3R compared to those in the control group (liver, 244 vs. 106; lungs, 103 vs. 28; kidney, 134 vs. 59, p<0.05) (Figure 5B and C). In addition, histopathological H&E staining of liver sections revealed that ginsenoside Rg3R significantly reduced the number of HT29-induced metastatic lesions and decreased the tissue density (Figure 5D). The above findings were consistent with the hypothesis that ginsenoside Rg3R suppresses metastasis in CRC cells in vivo using a mouse model of metastasis.
Discussion
Ginsenoside Rg3R and Rg3S are produced by epimerization at carbon-20 during the steaming process and are known to exert multiple biological effects.15 A previous study demonstrated that ginsenoside Rg3R can promote the storage of hepatic glycogen and decrease the accumulation of metabolites, such as lactic acid and serum urea nitrogen, ultimately reducing fatigue after intranasal administration.40 In addition, the antioxidant properties of ginsenoside Rg341 and its stimulatory effects on the immune response42 were found to be stereospecific. In particular, the R form of ginsenoside Rg3 exerts significantly stronger antioxidant effects and more adjuvant activity compared to the S form. Ginsenoside Rg3R has been shown to exert anti-cancer effects against melanoma,18 liver cancer,17 and lung cancer.16 Ginsenoside Rg3R can effectively inhibit the melanoma cell growth in vivo by downregulation of FUT436 or HDAC3,43 observed with a significant reduction of tumor size and volume. Similar with tumor growth in melanoma xenograft mouse model, ginsenoside Rg3R was shown to suppress the lung metastasis of melanoma B16-BL6 using a spontaneous metastasis model.18 Moreover, encapsulation of ginsenoside Rg3R using polylactic-co-glycolic acid (PLGA) was demonstrated to enhance its antitumor activity.44,45
In the present study, we examined the effects of ginsenoside Rg3R on CSC properties and the EMT in CRC cells. Results revealed that ginsenoside Rg3R significantly suppressed CSC characteristics and motility of CRC cells in vitro. Moreover, the number and size of metastasis nodules in liver, lung, and kidney tissues in a mouse metastasis model were decreased after ginsenoside Rg3R treatment. Mechanistically, EGFR/SNAIL signaling was significantly suppressed by ginsenoside Rg3R, consistent with previous studies demonstrating that ginsenosides, including ginsenoside Rg3, could strongly interact with EGFR;30 several ginsenosides, such as compound K31,32 and Rh2,35 are known to suppress EGFR signaling. In addition, our results revealed that the expression signatures of genes related to CSC properties and the EMT, including NANOG, SOX2, MMP2, and SNAIL, were dysregulated following treatment with ginsenoside Rg3R. NANOG and SOX2 are crucial CSC signatures because of their roles in self-renewal and maintenance of the undifferentiated state of CSCs.46,47 Besides, SNAIL, a potent transcriptional repressor of E-cadherin expression,48 has been previously implicated in the maintenance of CSC and the EMT. Furthermore, SNAIL has been associated with liver and lymph node metastasis, chemo-resistance, and poor prognosis because of its roles in transcriptional regulation.49 Thus, our current results, which demonstrated that ginsenoside Rg3R regulates the expression signatures of CSC and EMT-related genes, have very important implications in CRC treatment because these are critically implicated in cancer relapse and metastasis.
Two monoclonal antibodies targeting EGFR, namely, cetuximab and panitumumab, have been recently tested for their inhibitory effect on CRC. The clinical outcomes of metastatic CRC patients have been improved by the introduction of these two monoclonal antibodies in combination with chemotherapy.50 However, these drugs exerted mild toxic effects and were shown to be effective only in a small proportion of CRC patients, corresponding to clinically meaningful response rate of approximately 10.51,52 Given that ginsenoside Rg3R binds EGFR and regulates EGFR signaling, our findings suggested that ginsenoside Rg3R could be used alone or in combination with other drugs or antibodies that target EGFR to regulate EGFR signaling. However, there is a need to improve our understanding of the mechanisms underlying the anticancer activity of ginsenoside Rg3R against CRC. In particular, more detailed studies on ginsenoside Rg3R-EGFR interaction are required to investigate the binding between ginsenoside Rg3R and EGFR and to elucidate the mechanisms by which ginsenoside Rg3R regulates EGFR together with SNAIL. Ultimately, such studies can serve as the basis for the development of novel CRC treatments for the clinical management, in which ginsenoside Rg3R can be used as an essential inhibitory agent.
Acknowledgments
The present study was supported by a grant from the Korean Society of Ginseng funded by the Korea Ginseng Cooperation (2018) and the Soonchunhyang University Research Fund.
Disclosure
The authors declare no conflict of interest.
References
1. Misiakos EP, Karidis NP, Kouraklis G. Current treatment for colorectal liver metastases. World J Gastroenterol. 2011;17(36):4067–4075. doi:10.3748/wjg.v17.i36.4067
2. Mishra J, Drummond J, Quazi SH, et al. Prospective of colon cancer treatments and scope for combinatorial approach to enhanced cancer cell apoptosis. Crit Rev Oncol Hematol. 2013;86(3):232–250. doi:10.1016/j.critrevonc.2012.09.014
3. Ait Ouakrim D, Pizot C, Boniol M, et al. Trends in colorectal cancer mortality in Europe: retrospective analysis of the WHO mortality database. BMJ. 2015;351:h4970. doi:10.1136/bmj.h4970
4. Wahab SMR, Islam F, Gopalan V, Lam AK. The identifications and clinical implications of cancer stem cells in colorectal cancer. Clin Colorectal Cancer. 2017;16(2):93–102. doi:10.1016/j.clcc.2017.01.011
5. Cherciu I, Barbalan A, Pirici D, Margaritescu C, Saftoiu A. Stem cells, colorectal cancer and cancer stem cell markers correlations. Curr Health Sci J. 2014;40(3):153–161. doi:10.12865/CHSJ.40.03.01
6. Vaiopoulos AG, Kostakis ID, Koutsilieris M, Papavassiliou AG. Colorectal cancer stem cells. Stem Cells. 2012;30(3):363–371. doi:10.1002/stem.1031
7. Ren F, Sheng WQ, Du X. CD133: a cancer stem cells marker, is used in colorectal cancers. World J Gastroenterol. 2013;19(17):2603–2611. doi:10.3748/wjg.v19.i17.2603
8. Phi LTH, Sari IN, Yang YG, et al. Cancer stem cells (CSCs) in drug resistance and their therapeutic implications in cancer treatment. Stem Cells Int. 2018;2018:5416923. doi:10.1155/2018/5416923
9. Zhou Y, Xia L, Wang H, et al. Cancer stem cells in progression of colorectal cancer. Oncotarget. 2018;9(70):33403–33415. doi:10.18632/oncotarget.v9i70
10. Shin BK, Kwon SW, Park JH. Chemical diversity of ginseng saponins from panax ginseng. J Ginseng Res. 2015;39(4):287–298. doi:10.1016/j.jgr.2014.12.005
11. Nag SA, Qin JJ, Wang W, Wang MH, Wang H, Zhang R. Ginsenosides as anticancer agents: in vitro and in vivo activities, structure-activity relationships, and molecular mechanisms of action. Front Pharmacol. 2012;3:25. doi:10.3389/fphar.2012.00025
12. Li Y, Wang Y, Niu K, et al. Clinical benefit from EGFR-TKI plus ginsenoside Rg3 in patients with advanced non-small cell lung cancer harboring EGFR active mutation. Oncotarget. 2016;7(43):70535–70545. doi:10.18632/oncotarget.12059
13. Lee IS, Uh I, Kim KS, et al. Anti-inflammatory effects of ginsenoside Rg3 via NF-kappaB pathway in A549 cells and human asthmatic lung tissue. J Immunol Res. 2016;2016:7521601. doi:10.1155/2016/7521601
14. Lee B, Sur B, Park J, et al. Ginsenoside rg3 alleviates lipopolysaccharide-induced learning and memory impairments by anti-inflammatory activity in rats. Biomol Ther (Seoul). 2013;21(5):381–390. doi:10.4062/biomolther.2013.053
15. Kang KS, Kim HY, Yamabe N, Yokozawa T. Stereospecificity in hydroxyl radical scavenging activities of four ginsenosides produced by heat processing. Bioorg Med Chem Lett. 2006;16(19):5028–5031. doi:10.1016/j.bmcl.2006.07.071
16. Kim YJ, Choi WI, Jeon BN, et al. Stereospecific effects of ginsenoside 20-Rg3 inhibits TGF-beta1-induced epithelial-mesenchymal transition and suppresses lung cancer migration, invasion and anoikis resistance. Toxicology. 2014;322:23–33. doi:10.1016/j.tox.2014.04.002
17. Li X, Guan YS, Zhou XP, et al. Anticarcinogenic effect of 20(R)-ginsenoside Rg3 on induced hepatocellular carcinoma in rats. Sichuan Da Xue Xue Bao Yi Xue Ban. 2005;36(2):217–220.
18. Mochizuki M, Yoo YC, Matsuzawa K, et al. Inhibitory effect of tumor metastasis in mice by saponins, ginsenoside-Rb2, 20(R)- and 20(S)-ginsenoside-Rg3, of red ginseng. Biol Pharm Bull. 1995;18(9):1197–1202. doi:10.1248/bpb.18.1197
19. Fang Y, Eglen RM. Three-dimensional cell cultures in drug discovery and development. SLAS Discov. 2017;22(5):456–472. doi:10.1177/1087057117696795
20. Langhans SA. Three-dimensional in vitro cell culture models in drug discovery and drug repositioning. Front Pharmacol. 2018;9:6. doi:10.3389/fphar.2018.00006
21. Shoemaker RH, Wolpert-DeFilippes MK, Kern DH, et al. Application of a human tumor colony-forming assay to new drug screening. Cancer Res. 1985;45(5):2145–2153.
22. Rotem A, Janzer A, Izar B, et al. Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc Natl Acad Sci U S A. 2015;112(18):5708–5713. doi:10.1073/pnas.1505979112
23. Wylie PG, Bowen WP. Determination of cell colony formation in a high-content screening assay. Clin Lab Med. 2007;27(1):193–199. doi:10.1016/j.cll.2006.12.008
24. Podberezin M, Wen J, Chang CC. Cancer stem cells: a review of potential clinical applications. Arch Pathol Lab Med. 2013;137(8):1111–1116. doi:10.5858/arpa.2012-0494-RA
25. Stuelten CH, Parent CA, Montell DJ. Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat Rev Cancer. 2018;18(5):296–312. doi:10.1038/nrc.2018.15
26. Yilmaz M, Christofori G. Mechanisms of motility in metastasizing cells. Mol Cancer Res. 2010;8(5):629–642. doi:10.1158/1541-7786.MCR-10-0139
27. Spano JP, Lagorce C, Atlan D, et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol. 2005;16(1):102–108. doi:10.1093/annonc/mdi006
28. Italiano A, Saint-Paul MC, Caroli-Bosc FX, et al. Epidermal growth factor receptor (EGFR) status in primary colorectal tumors correlates with EGFR expression in related metastatic sites: biological and clinical implications. Ann Oncol. 2005;16(9):1503–1507. doi:10.1093/annonc/mdi282
29. Kluftinger AM, Robinson BW, Quenville NF, Finley RJ, Davis NL. Correlation of epidermal growth factor receptor and c-erbB2 oncogene product to known prognostic indicators of colorectal cancer. Surg Oncol. 1992;1(1):97–105. doi:10.1016/0960-7404(92)90062-P
30. Park K, Cho AE. Using reverse docking to identify potential targets for ginsenosides. J Ginseng Res. 2017;41(4):534–539. doi:10.1016/j.jgr.2016.10.005
31. Dougherty U, Mustafi R, Wang Y, et al. American ginseng suppresses Western diet-promoted tumorigenesis in model of inflammation-associated colon cancer: role of EGFR. BMC Complement Altern Med. 2011;11:111. doi:10.1186/1472-6882-11-111
32. Chen W, Qiu Y. Ginsenoside Rh2 targets EGFR by up-regulation of miR-491 to enhance anti-tumor activity in hepatitis B virus-related hepatocellular carcinoma. Cell Biochem Biophys. 2015;72(2):325–331. doi:10.1007/s12013-014-0456-9
33. Phi LTH, Wijaya YT, Sari IN, Yang YG, Lee YK, Kwon HY. The anti-metastatic effect of ginsenoside Rb2 in colorectal cancer in an EGFR/SOX2-dependent manner. Cancer Med. 2018;7(11):5621–5631. doi:10.1002/cam4.1800
34. Phi LTH, Sari IN, Wijaya YT, et al. Ginsenoside Rd inhibits the metastasis of colorectal cancer via epidermal growth factor receptor signaling axis. IUBMB Life. 2018.
35. Li S, Gao Y, Ma W, et al. EGFR signaling-dependent inhibition of glioblastoma growth by ginsenoside Rh2. Tumour Biol. 2014;35(6):5593–5598. doi:10.1007/s13277-014-1739-x
36. Shan X, Aziz F, Tian LL, Wang XQ, Yan Q, Liu JW. Ginsenoside Rg3-induced EGFR/MAPK pathway deactivation inhibits melanoma cell proliferation by decreasing FUT4/LeY expression. Int J Oncol. 2015;46(4):1667–1676. doi:10.3892/ijo.2015.2886
37. Joo EJ, Chun J, Ha YW, Ko HJ, Xu MY, Kim YS. Novel roles of ginsenoside Rg3 in apoptosis through downregulation of epidermal growth factor receptor. Chem Biol Interact. 2015;233:25–34. doi:10.1016/j.cbi.2015.03.016
38. Tian L, Shen D, Li X, et al. Ginsenoside Rg3 inhibits epithelial-mesenchymal transition (EMT) and invasion of lung cancer by down-regulating FUT4. Oncotarget. 2016;7(2):1619–1632. doi:10.18632/oncotarget.v7i2
39. Wang Y, Shi J, Chai K, Ying X, Zhou BP. The role of snail in EMT and tumorigenesis. Curr Cancer Drug Targets. 2013;13(9):963–972. doi:10.2174/15680096113136660102
40. Tang W, Zhang Y, Gao J, Ding X, Gao S. The anti-fatigue effect of 20(R)-ginsenoside Rg3 in mice by intranasally administration. Biol Pharm Bull. 2008;31(11):2024–2027. doi:10.1248/bpb.31.2024
41. Wei X, Su F, Su X, Hu T, Hu S. Stereospecific antioxidant effects of ginsenoside Rg3 on oxidative stress induced by cyclophosphamide in mice. Fitoterapia. 2012;83(4):636–642. doi:10.1016/j.fitote.2012.01.006
42. Wei X, Chen J, Su F, Su X, Hu T, Hu S. Stereospecificity of ginsenoside Rg3 in promotion of the immune response to ovalbumin in mice. Int Immunol. 2012;24(7):465–471. doi:10.1093/intimm/dxs043
43. Shan X, Fu YS, Aziz F, Wang XQ, Yan Q, Liu JW. Ginsenoside Rg3 inhibits melanoma cell proliferation through down-regulation of histone deacetylase 3 (HDAC3) and increase of p53 acetylation. PLoS One. 2014;9(12):e115401. doi:10.1371/journal.pone.0115401
44. Geng L, Fan J, Gao QL, Yu J, Hua BJ. Preliminary study for the roles and mechanisms of 20(R)-ginsenoside Rg3 and PEG-PLGA-Rg3 nanoparticles in the Lewis lung cancer mice. Beijing Da Xue Xue Bao Yi Xue Ban. 2016;48(3):496–501.
45. Zhang S, Liu J, Ge B, et al. Enhanced antitumor activity in A431 cells via encapsulation of 20(R)-ginsenoside Rg3 in PLGA nanoparticles. Drug Dev Ind Pharm. 2017;43(10):1734–1741. doi:10.1080/03639045.2017.1339079
46. You L, Guo X, Huang Y. Correlation of cancer stem-cell markers OCT4, SOX2, and NANOG with clinicopathological features and prognosis in operative patients with rectal cancer. Yonsei Med J. 2018;59(1):35–42. doi:10.3349/ymj.2018.59.1.35
47. Izumi D, Ishimoto T, Sakamoto Y, Miyamoto Y, Baba H. Molecular insights into colorectal cancer stem cell regulation by environmental factors. J Cancer Metastasis Treat. 2015;1(3):156–162. doi:10.4103/2394-4722.165532
48. Yook JI, Li XY, Ota I, Fearon ER, Weiss SJ. Wnt-dependent regulation of the E-cadherin repressor snail. J Biol Chem. 2005;280(12):11740–11748. doi:10.1074/jbc.M413878200
49. Cao H, Xu E, Liu H, Wan L, Lai M. Epithelial-mesenchymal transition in colorectal cancer metastasis: a system review. Pathol Res Pract. 2015;211(8):557–569. doi:10.1016/j.prp.2015.05.010
50. Leto SM, Trusolino L. Primary and acquired resistance to EGFR-targeted therapies in colorectal cancer: impact on future treatment strategies. J Mol Med (Berl). 2014;92(7):709–722. doi:10.1007/s00109-014-1161-2
51. Van Emburgh BO, Sartore-Bianchi A, Di Nicolantonio F, Siena S, Bardelli A. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol Oncol. 2014;8(6):1084–1094. doi:10.1016/j.molonc.2014.05.003
52. Cunningham D, Humblet Y, Siena S, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351(4):337–345. doi:10.1056/NEJMoa033025
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.