Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 10 » Issue 1
Variants in multiple genes polymorphism association analysis of COPD in the Chinese Li population
Authors Ding Y, Yang D, Zhou L, Xu J, Chen Y, He P, Yao J, Chen J, Niu H, Sun P, Jin T
Received 16 April 2015
Accepted for publication 13 May 2015
Published 27 July 2015 Volume 2015:10(1) Pages 1455—1463
DOI https://doi.org/10.2147/COPD.S86721
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Richard Russell
Yipeng Ding,1,* Danlei Yang,2,3,* Long Zhou,4 Junxu Xu,5 Yu Chen,5 Ping He,1 Jinjian Yao,1 Jiannan Chen,1 Huan Niu,1 Pei Sun,1 Tianbo Jin4
1Department of Emergency, People’s Hospital of Hainan Province, Haikou, Hainan, 2Department of Respiratory and Critical Care Medicine, Tongji Hospital, Key Laboratory of Pulmonary Diseases of Health Ministry, Tongji Medical College, 3Department of Science and Technology, Huazhong University, Wuhan, 4School of Life Sciences, Northwest University, Xi’an, 5Department of Respiration Emergency, The Third People’s Hospital of Haikou, Haikou, Hainan, People’s Republic of China
*These authors contributed equally to this work
Background: It is known that the contribution of risk alleles to chronic obstructive pulmonary disease (COPD) may vary between populations. Further, previous studies involving various ethnic groups have revealed associations between COPD and genetic polymorphisms in families with sequence similarity 13, member A (FAM13A), micro-RNA 2054 (MIR2054), SET domain containing protein 7 (SETD7), ring finger protein 150 (RNF150), hedgehog interacting protein (HHIP), and vascular endothelial growth factor A (VEGFA). Our objective was to explore the association between these gene polymorphism and COPD in members of Chinese Li minority population.
Materials and methods: The Chinese Li population case–control study was conducted to assess genetic associations with COPD risk. Seven single nucleotide polymorphisms (SNPs) located on chromosome 4, including FAM13A, MIR2054, SETD7, RNF150, and HHIP, and nine SNPs in the VEGFA gene were genotyped among 234 cases and 240 controls using Sequenom Mass-ARRAY® platform. Linkage disequilibrium (LD) analysis was performed using Haploview software and the associations of the SNP frequencies with COPD were analyzed using chi-square (χ2) tests, genetic models analysis, and haplotype analysis.
Results: By χ2 we found the minor allele “G” of rs17050782 was with increased COPD risk in allele model. In genetic models, we found the minor allele of rs7671167 (P=0.028 by dominant model) and rs17050782 (P=0.008 by recessive model) was associated with the increased risk of COPD disease. Likewise, an increased risk of developing COPD was associated with the “GGCGC” haplotype of VEGFA (odds ratio =1.48, 95% confidence interval =1.02–2.12, P=0.037).
Conclusion: Our results were the first time to reveal that SNPs from FAM13A (rs7671167), SETD7 (rs17050782), and a haplotype of VEGFA (“GGCGC”) are potential susceptibility loci associated with increased COPD risk in Chinese Li minority population.
Keywords: chronic obstructive pulmonary disease, case–control study, single nucleotide polymorphism, Chinese Li minority population
Introduction
The Li population, which is one among the 55 minority ethnic groups in the People’s Republic of China, exceeds 1.3 million and resides primarily in the Li and Miao Autonomous Prefecture in the center and southwest regions of the Hainan Province. However, a small number of the Li population members are intermixed with the Han people. Among the male Li villagers, the prevalence rates of tobacco users (84.1% were lifetime smokers) and (92.5% were lifetime drinkers) are higher than those of the national average and other ethnic minorities in People’s Republic of China, but fewer Li females reported smoking (1.9% were lifetime smokers) or alcohol consumption (21.0% were lifetime drinkers).1
Chronic obstructive pulmonary disease (COPD) is defined as airflow limitation that is not completely reversible. COPD is typically caused by exposure to noxious particles or gases, predominantly cigarette smoking; however, other exposures such as biomass fuels are an important cause worldwide.2 In fact, COPD is one of the leading causes of morbidity and mortality worldwide3,4 and is expected to be the third leading cause of worldwide mortality and the fifth leading cause of morbidity by the year 2020.4
Both genetic and environmental factors contribute to the disease etiology of COPD, and substantial evidence suggests that a genetic susceptibility to COPD exists.5 Recently, the application of genome-wide association studies (GWAS) has enabled the identification of a number of common variants involved in the etiology of COPD. In large studies including subjects with a range of smoking histories and damaged lung function, GWAS results have revealed genomic regions that demonstrate a highly and reproducible association with COPD. The regions are located in family with sequence similarity 13, member A (FAM13A) on chromosome 4q22,6 and near hedgehog interacting protein (HHIP) on chromosome 4q31.7 One prior GWAS involving 2,940 cases and 1,380 smoking controls with normal lung function identified common genetic risk variants in FAM13A [rs7671167, odds ratio (OR) =0.76, combined P=1.2×10−11], and provided evidence of replication in one case–control and two family-based cohorts in Norway.6 Other studies of HHIP, which encode an inhibitory protein for sonic hedgehog, revealed that single nucleotide polymorphisms (SNPs) upstream of HHIP both modulate the expression of HHIP and functionally link reduced HHIP expression to COPD pathogenesis,8 and are crucial for the development of the lungs and other organs.9 Additional studies identified three SNPs on HHIP (rs13118928, rs13141641, and rs1828591) that were associated with COPD and lung function in non-Asian populations.7,10,11 Recently, the association of HHIP polymorphisms with COPD and COPD-related phenotypes has been found in Chinese Han population.3 A separate association study suggested that the genetic predisposition to COPD and to lung cancer could share common pathogenetic factors including the 4q22.1 locus, which implicating the Rho-kinase pathway.12 It is well known that smoking is associated with lung cancer; thus, we selected candidate SNPs (rs950063, rs17050782, and rs10007052) between FAM13A and HHIP from chromosome 4 that are reportedly associated with cigarette smoking behaviors.13,14 Vascular endothelial growth factor A (VEGFA), which is located on chromosome 6p21.1, is an important gene that encodes protein with potent angiogenic properties that enhances vascular permeability and modulates thrombogenicity. The biological properties of VEGFA have led to interest in its role within the lung in both healthy and diseased states.15 In fact, studies involving lung tissue obtained from patients with COPD have suggested that the expression of VEGFA may play a role in the pathogenesis of the disease.16,17 As well, a previous study suggested that VEGFA may be an important factor in both chronic lung and cardiovascular disease processes, and could link COPD with cardiovascular disorders.18
Previous COPD association studies have focused on the Chinese Han nationality and other large groups, but specific data on the Li population are lacking. Because of the various genetic backgrounds among different nationalities, one aim of this study was to verify whether COPD susceptibility loci from VEGFA and the genetic risk variants in FAM13A and HHIP identified in prior GWAS were significant in the Chinese Li minority population. The second objective was to try to find new FAM13A and HHIP susceptibility loci for COPD among smoking behaviors in Chinese Li minority individuals. To this end, we anticipated that we could successfully identify suitable and accurate molecular markers of COPD among the Chinese Li minority population.
Materials and methods
Study participants
The Chinese Li minority population-based case–control study comprising COPD patients diagnosed from January 2010 to December 2013 from the Hainan Province People’s Hospital was conducted. All subjects were Chinese Li minority individuals identified from the ID card information. All cases were diagnosed by a physician based on a COPD pulmonary function test (PFT) indicating a post-bronchodilator (post-BD) forced expiratory volume in 1 second (FEV1)/forced vital capacity (FVC) of less than 70% and the FEV1 of less than 80% predicted, and disease severity was classified according to the global initiative for chronic obstructive lung disease criteria.19 Patients were excluded from the study if they had an established diagnosis of asthma, lung cancer, a history of atopy, or a known α1-antitrypsin deficiency before PFT.3 A total of 1,734 subjects from the department of respiratory underwent PFT during this study period at the Hainan Province People’s Hospital. We also excluded 50 patients whose age was <40 and who did not complete the questionnaires. Among the remaining 1,684 patients, 421 had a pre-BD FEV1/FVC of <0.7. We excluded 153 patients who did not undergo post-BD spirometry. Among the remaining 268 patients, 34 had a post-BD FEV1/FVC of ≥0.7 and FEV1 of more than 80% predicted. Eventually, 234 patients consisted of 142 males and 92 females were found to have COPD with a post-BD FEV1/FVC of <0.7 (Figure 1). Control subjects were randomly recruited from the health centers of Hainan Province Hospital during the same period. Exclusion criteria were as described for cases and additionally included family history of COPD. Finally, 240 controls consisting of 151 males and 89 females with a mean age of 62.09 years, who were at least 40 years old, and in good mental health were recruited in this study. Detailed population information for the study participants is shown in Table 1.
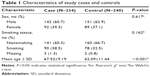  | Table 1 Characteristics of study cases and controls |
Spirometry and calculated method
COPD is defined as a post-BD FEV1/FVC ratio of <0.7 in patients aged ≥40 years, according to the Global Initiative for Chronic Obstructive Lung Disease (GOLD) guideline.20 Spirometry results were expressed as percentages of the predicted values and were calculated using Morris’s predictive equations as follows: predicted FEV1 for men =0.092× height −0.032× age −1.26, predicted FEV1 for women =0.089× height −0.025× age −1.932 (height in inches).21
Spirometry was performed using spirometers (models: VMAX229, VMAX22, and VMAX20; SensorMedics, Yorba Linda, CA, USA) by trained technicians according to the 2005 American Thoracic Society/European Respiratory Society recommendations.22,23
Demographic and clinical data
To obtain the basic research data, demographic and personal information were collected through the use of a standardized questionnaire that included smoking history status (we defined never smokers as individuals who had smoked less than 100 cigarettes during their lifetime), weight, body mass index [BMI = weight (kg)/height (m2)], and special examinations including electrocardiogram, pulmonary function, and color sonography. Each procedure was conducted by the same professionals, and the questionnaire survey staff was methodically trained to ensure strict quality control of the administration of the questionnaire during the investigation, and ensured all participants signed on the questionnaire. The case information was collected through consultation with the treating physicians or by reviewing the medical chart. Written informed consent was obtained from all subjects, and the study was approved by the ethics committee of Hainan Province People’s Hospital.
SNP selection and genotyping
Targeted reuse of existing GWAS data is a reported approach for identifying additional putative risk alleles.24 Accordingly, one SNP (rs7671167) was chosen from FAM13A and three SNPs (rs1828591, rs13118928, and rs13141641) were chosen from HHIP. All of the selected SNPs were in the region of chromosome 4q and were associated with COPD and COPD-related phenotypes in non-Asian populations according to recent GWAS,6,7,10,11 and were subsequently verified in Chinese Han populations.3 Three SNPs (rs950063, rs17050782, and rs10007052) located in the region of chromosome 4q between FAM13A and HHIP were all in the region of chromosome 4q and were selected based on reported associations with cigarette smoking behaviors.13,14 Nine SNPs (rs25648, rs833068, rs833070, rs3024994, rs3024997, rs3025000, rs3025030, rs3025033, and rs10434) located in VEGFA in the region of chromosome 6p21.1 that indicated both chronic lung and cardiovascular disease processes were selected, because VEGFA may be an important factor linking COPD with cardiovascular disease.17,18 Ultimately, we obtained 16 SNPs in different genes that were in Hardy–Weinberg equilibrium (HWE; P-value >0.05) in the control individuals. We extracted genomic DNA from peripheral blood using a GoldMag® – Mini Whole Blood Genomic DNA Purification Kit (GoldMag® Ltd., Xi’an, People’s Republic of China) according to the manufacturer’s protocol, and DNA concentrations were measured using a NanoDrop™ 2000C (Thermo Fisher Scientific, Waltham, MA, USA). We used the Sequenom Mass-ARRAY® Assay Design 3.0 software (Sequenom, Inc., San Diego, CA, USA) to design Multiplexed SNP Mass-EXTEND assays.25 SNP genotyping was performed using the standard protocol recommended by the manufacturer with a Sequenom Mass-ARRAY® RS1000 (Sequenom, Inc.). Sequenom Typer 4.0 software was used for data management and analyses.26
Statistical analyses
We used the SPSS 21.0 statistical packages (SPSS Inc., Chicago, IL, USA) and Microsoft Excel for statistical analysis. All P-values in this study were two sided, and P=0.05 was considered the threshold for statistical significance. The validation of each SNP frequency in control subjects was tested for departure from HWE using an exact test. Allele frequencies and genotype frequencies for each SNP of COPD patients and control subjects were compared using a chi-squared (χ2) test.27 Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated using unconditional logistic-regression analysis with adjustments for age, sex, and smoking status.28 We evaluated whether an interaction existed between covariate smoking status (smoking or non-smoking) and each SNP loci using logistic-regression models (recessive, dominant, and codominant models). We calculated the linkage disequilibrium (LD) coefficients and constructed haplotype using the Haploview software package (version 4.2) (Mark Daly’s Laboratory, Massachusetts Institute of Technology/Harvard Broad Institute, Cambridge, MA, USA).29 We used SNP Stats (Catalan Institute of Oncology, Barcelona, Spain), a web-based software to test the associations between certain SNPs and the risk of COPD in five genetic models (dominant, codominant, overdominant, recessive, and log-additive).30
Results
A total of 474 Li participants, including 234 COPD cases and 240 controls, were successfully genotyped for further analysis. Males represented 62.9% of the controls and 60.7% of the cases. Fewer females than males participated in this study. However, sex was equally distributed among COPD cases and control subjects. There was a significant difference between the cases and controls in terms of mean age distribution (67.52 in cases vs 62.09 in controls P<0.001; Table 1). Table 2 summarizes the basic characteristics of the SNPs in the study population, and shows that all 16 SNPs conformed to HWE in the controls (P>0.05). We compared the differences in frequency distributions of alleles between cases and controls by Pearson χ2 test and found only the minor allele “G” in rs17050782 from SET domain containing protein 7 (SETD7) was significantly associated with risk of COPD in the study population (P=0.014, OR=1.40, 95% CI=1.07–1.83).
We also used SNP Stats to assess the association between these SNPs and COPD risks using five genetic models (codominant, dominant, recessive, overdominant, and log-additive) by unconditional logistic-regression analysis. As shown in Table 3, crude analysis revealed the genotype “G/G” in rs17050782 was associated with increased risk of COPD under the codominant model (P=0.007, OR=2.64, 95% CI=1.40–5.00), under the recessive model (P=0.002, OR=2.52, 95% CI=1.38–4.61), and under log-additive model (P=0.012, OR=1.42, 95% CI=1.08–1.87). After adjustment for sex, age, and smoking status, we found two SNPs, rs7671167 located in FAM13A and rs17050782 located in SETD7, were associated with increased COPD risk. The genotypes “T/C” and “T/T” in rs7671167 from FAM13A were associated with increased COPD risk (P=0.028, OR=1.58, 95% CI=1.05–2.38) under the dominant model, whereas the genotype “G/G” in rs17050782 from SETD7 was associated with increased COPD risk under the codominant model (P=0.026, OR=2.41, 95% CI=1.24–4.69), under the recessive model (P=0.008, OR=2.30, 95% CI=1.22–4.31), and under the log-additive model (P=0.03, OR=1.38, 95% CI=1.03–1.85) (Table 4). Perhaps due to the genetic differences between the Li population and other large populations or because of the small sample size, we did not identify any associations between SNPs located in HHIP and COPD in Chinese Li minority population included in this study.
One haplotype block that included two SNPs (rs1828591 and rs13118928) with D′=1 (Figure 2) was detected in HHIP SNPs by haplotype analyses. Haplotypes with frequencies >1% were selected for analysis, and the association between inferred haplotypes and COPD risk among the individuals was analyzed. However, we found no association between the selected haplotype and COPD risk. Two haplotype blocks that included five SNPs (rs833068, rs833070, rs3024994, rs3024997, and rs3025000) with D′=1 (Figure 3) were detected in VEGFA SNPs by haplotype analyses. The five SNPs constructed four haplotypes (“AGCAT”, “GACGC”, “GGCGC”, and “GGTGC”), and the haplotype “GGCGC” in the VEGFA gene was significantly associated with increased risk of COPD under the logistic-regression model adjusted for sex, age, and smoking status (OR=1.48, 95% CI=1.02–2.12, P=0.037). The results of the association between the VEGFA haplotype and the risk of COPD are listed in Table 5. Another haplotype block that included two SNPs (rs3025030 and rs3025033) is shown in Figure 3, but the haplotypes were not found to be associated with COPD risk.
Discussion
In this case–control study that included an ample number of COPD cases and controls (234 cases and 240 controls) from the Chinese Li minority population, we determined that the rs7671167 SNP from FAM13A (adjusted P-value =0.028 under the dominant model) and the rs17050782 SNP from SETD7 (adjusted P-value =0.008 under the recessive model) were associated with an increased risk of COPD. Likewise, we observed that the haplotype “GGCGC” in VEGFA was associated with an increased risk of developing COPD (OR=1.48, 95% CI=1.02–2.12, P=0.037) under the logistic-regression model adjusted for sex, age, and smoking status.
COPD is an escalating global health problem caused by multiple genetic and environmental factors.5 Further, even if the same genetic variant was involved in each population, the LD relationships of this variant with neighboring genetic polymorphisms would likely vary between ethnic groups. The rs7671167 SNP, which was first described by Cho et al, is located in intron 4 of FAM13A and is in LD with SNPs in the Rho-GTPase activating proteins domain region.6 Previous studies have confirmed that in Caucasians, the FAM13A rs7671167 variant confers a protective effect on smoking-related COPD alone (C allele, OR=0.79, P=0.013; and CC genotype, OR=0.71, P=0.024) and on COPD, both with and without lung cancer (C allele, OR=0.80, P=0.008; and CC genotype, OR=0.70, P=0.007).12 In this study, we also confirmed that the association of the rs7671167 loci with COPD, but we studied a different ethnic group; our results indicated that the minor allele “T” in rs7671167 was associated with increased COPD risk (P=0.028, OR=1.58, 95% CI=1.05–2.38) under the dominant model (T/C, T/T, vs CC).
To date, the biological function of the rs7671167 SNP located in FAM13A has not been fully characterized.6 However, prior studies have reported that FAM13A is an interesting candidate gene because of the Rho-GTPase activating proteins domain it encodes,31 and because of its associated tumor suppressor activity via inhibition of the intracellular signal transduction molecule Rho-A.32 In the present study, we also discovered a new SNP (rs17050782) located in SETD7 code super antigen-like protein 5 that was associated with increased risk of COPD under the recessive model (P=0.008, OR=2.30, 95% CI=1.22–4.31). Previous study mentioned that the SNP (rs17050782) was predicted with cigarette smoking behaviors from the GWAS (P<10−5) but did not verified coding regions associated with cigarette smoking behaviors.14 VEGF is a potent mediator of angiogenesis that has multiple effects on lung development and physiology. As well, previous studies have suggested the up-regulation of systemic inflammation and circulating VEGF levels in patients with acute exacerbated COPD.18 Although our findings indicated that no associations existed between SNPs in VEGFA and COPD, which was not in accord with the results of previous studies, we observed that the haplotype “GGCGC” in VEGFA was associated with increased risk of COPD under the logistic-regression model adjusted for sex, age, and smoking status (OR=1.48, 95% CI=1.02–2.12, P=0.037).
The present study had several limitations. First, despite our ability to identify genetic associations with COPD, we were not able to elucidate causal mechanisms. Nonetheless, despite the limitations of our small sample size, we were still able to demonstrate several associations with COPD.33 However, a larger sample size and a greater number of loci might have allowed for additional evidence regarding the role of these genes in COPD. Second, we did not account for the effects of secondhand smoke or other smoke in the “no smoking” individuals. Hence, our future research should incorporate the analysis of the relationship between COPD and detailed smoking status including smoking intensity, secondhand smoking, and air smog. Finally, considering the marked difference in smoking prevalence among sex, which is an established risk factor for COPD, sex differences could exist. However, because the sample size was not large enough, we did not attempt separate analysis for males or females and other stratified analyses. However, if analyzed in-depth, these study data could provide important clues regarding COPD pathogenesis, especially given the fact that we determined the rs7671167 SNP with “T” allele was associated with enhanced risk of COPD, in contrast to the already published study that reported the rs7671167 SNP with a “C” allele demonstrated a protective effect.
In summary, we are the first to identify two candidate loci (rs7671167 and rs17050782) associated with increased risk of COPD in the Chinese Li minority population. As well, we are the first to report that the “GGCGC” haplotype in the VEGFA was associated with an increased risk of developing COPD. However, we are aware that prior to using these loci and haplotype as molecular markers for the detection and prevention of COPD, further studies are required to elucidate the functional variants that confer a risk for COPD in the associated genomic regions.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (No 81160008). We are grateful to the COPD patients and control subjects for their participation in this study. We also thank the clinicians and hospital staffs who contributed to the sample and data collection.
Disclosure
The authors report no conflicts of interest in this work.
References
Tang X, Chen X, Li H, Stanton B, Li X. Smoking and drinking patterns among residents of Li ethnic minority villages in Hainan, China. Subst Use Misuse. 2005;40(5):687–701. | ||
Rabe KF, Hurd S, Anzueto A, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176(6):532–555. | ||
Wang B, Zhou H, Yang J, et al. Association of HHIP polymorphisms with COPD and COPD-related phenotypes in a Chinese Han population. Gene. 2013;531(1):101–105. | ||
Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. | ||
Mannino DM, Buist AS. Global burden of COPD: risk factors, prevalence, and future trends. Lancet. 2007;370(9589):765–773. | ||
Cho MH, Boutaoui N, Klanderman BJ, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42(3):200–202. | ||
Wilk JB, Chen TH, Gottlieb DJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5(3):e1000429. | ||
Zhou X, Baron RM, Hardin M, et al. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Hum Mol Genet. 2012;21(6):1325–1335. | ||
Zhou X, Qiu W, Sathirapongsasuti JF, et al. Gene expression analysis uncovers novel hedgehog interacting protein (HHIP) effects in human bronchial epithelial cells. Genomics. 2013;101(5):263–272. | ||
Pillai SG, Ge D, Zhu G, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5(3):e1000421. | ||
Cho MH, Castaldi PJ, Wan ES, et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum Mol Genet. 2012;21(4):947–957. | ||
Young RP, Hopkins RJ, Hay BA, Whittington CF, Epton MJ, Gamble GD. FAM13A locus in COPD is independently associated with lung cancer – evidence of a molecular genetic link between COPD and lung cancer. Appl Clin Genet. 2011;4:1–10. | ||
Siedlinski M, Cho MH, Bakke P, et al. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66(10):894–902. | ||
Caporaso N, Gu F, Chatterjee N, et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One. 2009;4(2):e4653. | ||
Kaner RJ, Crystal RG. Compartmentalization of vascular endothelial growth factor to the epithelial surface of the human lung. Mol Med. 2001;7(4):240–246. | ||
Kasahara Y, Tuder RM, Cool CD, Lynch DA, Flores SC, Voelkel NF. Endothelial cell death and decreased expression of vascular endothelial growth factor and vascular endothelial growth factor receptor 2 in emphysema. Am J Respir Crit Care Med. 2001;163(3 Pt 1):737–744. | ||
Kranenburg AR, de Boer WI, Alagappan VK, Sterk PJ, Sharma HS. Enhanced bronchial expression of vascular endothelial growth factor and receptors (Flk-1 and Flt-1) in patients with chronic obstructive pulmonary disease. Thorax. 2005;60(2):106–113. | ||
Valipour A, Schreder M, Wolzt M, et al. Circulating vascular endothelial growth factor and systemic inflammatory markers in patients with stable and exacerbated chronic obstructive pulmonary disease. Clin Sci (Lond). 2008;115(7):225–232. | ||
Pauwels RA, Buist AS, Calverley PM, Jenkins CR, Hurd SS; GOLD Scientific Committee. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease. NHLBI/WHO Global Initiative for Chronic Obstructive Lung Disease (GOLD) Workshop summary. Am J Respir Crit Care Med. 2001;163(5):1256–1276. | ||
Vestbo J, Hurd SS, Agusti AG, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2013;187(4):347–365. | ||
Morris JF, Koski A, Johnson LC. Spirometric standards for healthy nonsmoking adults. Am Rev Respir Dis. 1971;103(1):57–67. | ||
Miller MR, Hankinson J, Brusasco V, et al. Standardisation of spirometry. Eur Respir J. 2005;26(2):319–338. | ||
Choi SM, Lee J, Park YS, et al. Prevalence and global initiative for chronic obstructive lung disease group distribution of chronic obstructive pulmonary disease detected by preoperative pulmonary function test. PLoS One. 2015;10(1):e0115787. | ||
Buas MF, Levine DM, Makar KW, et al. Integrative post-genome-wide association analysis of CDKN2A and TP53 SNPs and risk of esophageal adenocarcinoma. Carcinogenesis. 2014;35(12):2740–2747. | ||
Gabriel S, Ziaugra L, Tabbaa D. SNP genotyping using the Sequenom MassARRAY iPLEX platform. Curr Protoc Hum Genet. 2009;Chapter 2:Unit 2.12. | ||
Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39(3):347–351. | ||
Adamec C. Example of the use of the nonparametric test. Test X2 for comparison of 2 independent examples. Cesk Zdrav. 1964;12:613–619. | ||
Bland JM, Altman DG. Statistics notes. The odds ratio. BMJ. 2000;320(7247):1468. | ||
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. | ||
Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. | ||
Cohen M, Reichenstein M, Everts-van der Wind A, et al. Cloning and characterization of FAM13A1 – a gene near a milk protein QTL on BTA6: evidence for population-wide linkage disequilibrium in Israeli Holsteins. Genomics. 2004;84(2):374–383. | ||
Ridley AJ. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11(12):471–477. | ||
Hardin M, Zielinski J, Wan ES, et al. CHRNA3/5, IREB2, and ADCY2 are associated with severe chronic obstructive pulmonary disease in Poland. Am J Respir Cell Mol Biol. 2012;47(2):203–208. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







