Back to Archived Journals » Advances in Genomics and Genetics » Volume 4
Two different BRCA2 mutations found in a multigenerational family with a history of breast, prostate, and lung cancers
Authors Caporale D, Swenson E
Received 3 March 2014
Accepted for publication 23 April 2014
Published 20 June 2014 Volume 2014:4 Pages 87—94
DOI https://doi.org/10.2147/AGG.S63411
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Diane A Caporale, Erica E Swenson
Department of Biology, University of Wisconsin – Stevens Point, Stevens Point, WI, USA
Abstract: Breast and lung cancer are two of the most common malignancies in the United States, causing approximately 40,000 and 160,000 deaths each year, respectively. Over 80% of hereditary breast cancer cases are due to mutations in two breast cancer predisposition genes, BRCA1 and BRCA2. These are tumor-suppressor genes associated with DNA repair. Since the discovery of these two genes in the mid-1990s, several other breast cancer predisposition genes have been identified, such as the CHEK2 gene encoding a regulator of BRCA1. Recently, studies have begun investigating the roles of BRCA1 and BRCA2 gene expression in lung cancer. We conducted a family-based case study that included a bloodline of Italian heritage with several cases of breast cancer and associated cancers (prostate and stomach) through multiple generations and on a nonblood relative of Scottish/Irish descent who was consecutively diagnosed with breast and lung cancer. Cancer history and environmental risk factors were recorded for each family member. To investigate possible genetic risks, we screened for mutations in specific hypervariable regions of the BRCA1, BRCA2, and CHEK2 genes. DNA was extracted and isolated from the individuals' hair follicles and cheek cells. Polymerase chain reaction (PCR), allele-specific PCR, and DNA sequencing were performed to identify and verify the presence or absence of mutations in these regions. Genotypes of several family members were determined and carriers of mutations were identified. Here we report for the first time the occurrence of two different BRCA2 frameshift mutations within the same family. Specifically, three Italian family members were found to be carriers of the BRCA2-c.2808_2811delACAA (3036delACAA) mutation, a 4-nucleotide deletion in exon 11, which is a truncated mutation that causes deleterious function of BRCA2. This mutation that has been reported in many women of Spanish descent is within a hotspot and is predicted to have resulted from three separate mutational events. Although sporadic mutations can occur, more than likely it is the result of a germ line mutation inherited from the Italian family line and was carried by a father that died of prostate cancer. Since individual III-2 had an early onset of breast cancer, it is recommended that siblings of II-1 seek genetic counseling and be screened for the BRCA2-3036delACAA variant. The individual with breast and lung cancer (II-8) was not a carrier of this mutation, but rather a carrier of the BRCA2-c.6275_6276delTT (6503delTT), which is also a truncated mutation but more common in those of Irish/Scottish descent. It is recommended that her immediate family members be screened for this mutation to assess their risk of breast cancer. We conclude that DNA screening of the BRCA2 promoter region and the BRCA2-6503delTT site from a lung tumor biopsy taken from individual II-8 would provide more insight into the possible association of this BRCA2 variant with lung cancer.
Keywords: breast/prostate/lung cancers, BRCA2 deletions, AS PCR, genogram
Introduction
The two most common malignancies in the United States are breast cancer and lung cancer, which cause nearly 40,000 and 160,000 deaths each year, respectively.1 Up to 10% of breast cancer cases are hereditary, with over 80% of those cases due to mutations in two breast cancer predisposition genes, BRCA1 and BRCA2.2,3 Researchers have established a strong association between numerous mutations in the BRCA1 and BRCA2 genes and the risk of developing breast cancer as well as ovarian, pancreatic, prostate, and stomach/pancreatic cancers.4 Recently, studies have begun investigating the role of epigenetics with varying BRCA1 and BRCA2 expression in non-small cell lung cancer.5–9
The BRCA1 and BRCA2 genes, located on chromosome 17 and 13, respectively, are tumor suppressor genes encoding large proteins involved in the TP53 signal transduction pathway that repairs double-strand breaks in DNA.8,10 Mutations of these genes can result in non- or dysfunctional proteins, leading to tumor growth and cancer. As listed in the Breast Cancer Information Core (BIC) database, clinically important BRCA1 mutations are often frameshift mutations, resulting in a truncated nonfunctional protein. The mutation has an inheritance pattern of autosomal dominant, which predisposes one to earlier onset of breast tumors.11 In addition, the incidence of BRCA1 varies between different populations, suggesting it interacts with other proteins and environmental factors. A large number of studies have looked at variants of the BRCA1 and BRCA2 genes to determine if a specific mutation is of clinical significance. By 2010, over 750 clinically important mutations have been identified in BRCA2.2 Because of the sheer number of mutations, many studies have scanned the entirety of the BRCA2 gene using sequence analysis to identify mutations. In addition, several researchers have examined how BRCA1/2 promoter hypermethylation, which reduces levels of BRCA1/2 messenger RNA, is related to lung cancer tumor growth.8
Since the discovery of the BRCA1 and BRCA2 genes in 1994 and 1995, respectively, several other breast cancer predisposition genes have been identified.12 The CHEK2 gene, located on chromosome 22, encodes DNA replication checkpoint kinase 2, which is an upstream regulator of BRCA1 and TP53. As expected, mutations of the CHEK2 gene have also been associated with a variety of cancers. Consequently, researchers have analyzed specific variants to determine the association between specific mutations and the risk of different types of cancer.13 A strong association between several mutations in the BRCA1/2 and CHEK2 genes has been established with the risk of developing breast cancer as well as ovarian, prostate, pancreatic, stomach, and colon cancer.4 In addition, CHEK2 mutations have been frequently found in women with hereditary breast cancer who tested negative for BRCA1 and BRCA2 mutations.14
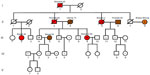
As illustrated in the pedigree in Figure 1, individual III-9 (the ninth individual from the third generation) of Italian descent is a 6-year breast cancer survivor whose breast cancer recently recurred. Her deceased father had prostate cancer, his sister had breast cancer twice, and his father had stomach cancer. A nonblood relative of Scottish/Irish decent, individual II-8 is a breast cancer survivor who was most recently diagnosed with non-small cell lung cancer. Individual III-9 received genetic testing by Myriad Genetics (Salt Lake City, Utah, USA) and tested positive for the BRCA2-c.2808_2811delACAA (3036delACAA) mutation. This frameshift mutation results in an early stop codon at amino acid position 958 of the BRCA2 protein, thus rendering it nonfunctional. We investigated the inheritance of the BRCA2-3036delACAA mutation throughout her extended family to assess their risk of developing breast cancer and other associated cancers, such as prostate and stomach cancer identified within the family. Participants were also screened for two variants of the CHEK2 gene, the (I157T) missense variant and (1100delC) truncating variant. Although mutations in CHEK2 are rarer than BRCA mutations, CHEK2 founder mutations from eastern Europe were also included in the study based on their association with an increased risk of breast, prostate, and stomach cancer, which were experienced in this extended family.13 In addition, we screened II-8’s DNA for possible BRCA1 or BRCA2 mutations typically found in Scottish and Irish populations to help provide insight into a possible genetic cause for her breast and lung cancers.15–17
Materials and methods
DNA isolation
DNA was isolated using the Qiagen (Valencia, CA, USA) DNeasy Kit. At least ten hair follicles from each individual were first washed in 100% ethanol and then placed in ATL lysis buffer and 30 μL of proteinase K. Samples were incubated at 55°C overnight. DNA was purified using spin columns and following the manufacturer’s instructions. Finally, products were eluted with 30 μL of Buffer AE. DNA concentrations were determined spectrophotometrically using a Thermo Scientific Nanodrop 1000 (Thermo Fisher Scientific, Waltham, MA, USA) and ranged from 50 ng/μL to 150 ng/μL.
Polymerase chain reaction and DNA sequencing
A 257-base pair (bp) portion of exon 11 of the BRCA2 gene was amplified using primers that were designed to flank the (3,036–3,039)-nucleotide region, where the 4-bp deletion would be located. National Center for Biotechnology Information GenBank accession # NM_000059.3 was used as a reference sequence of the BRCA2 gene, with the amplicon corresponding to region 26,613–26,869. The forward primer was BRCA2-3036del4-f: (5′-GACTTGACTTGTGTAAACGAACCC-3′), whereas the reverse primer was BRCA2-3036del4-r: (5′-CCTAAGAGTCCTGCCCATTTGTTC-3′). Between 50 ng and 100 ng of DNA was used in each polymerase chain reaction (PCR), which included a final concentration of 1× GoTaq® buffer (Promega Corporation, Fitchburg, WI, USA), 1.5 mM MgCl2, 0.2 mM of each deoxynucleoside triphosphate, 1 U of GoTaq® polymerase (Promega), 1 μM of each forward and reverse primer, and enough deionized water to reach a final volume of 25 μL. PCRs were performed in an MJ Research PTC100 thermocycler (Waltham, MA, USA). Samples were denatured at 94°C for 5 minutes, followed by 94°C for 30 seconds, annealed at 60°C for 60 seconds, and extended at 72°C for 120 seconds for 40 cycles, and ended with a final extension at 72°C for 5 minutes.
The CHEK2 gene was also screened for two founder mutations, (p.I157T) and (p.1100delC), using primer pairs as described by Cybulski et al.13 PCR conditions were the same as previously described, except the annealing temperature was 50°C. To verify amplification, 10 μL of each sample, a negative control, and a positive control, with each containing 2 μL of Orange G loading dye (NewEngland BioLabs, Ipswich, MA, USA), were separated by electrophoresis on a 1% Bio-Rad (Hercules, CA, USA) agarose gel containing ethidium bromide and 1× TAE (Tris acetate-ethylenediaminetetraacetic acid) and visualized on a BioRad transilluminator (Hercules, CA, USA). The remaining 15 μL of PCR product was then purified using Diffinity RapidTips (Sigma-Aldrich, St Louis, MO, USA). Samples were cycle-sequenced using Taq polymerase-mediated incorporation of dye-labeled dideoxy terminators. Sequencing reactions contained approximately 50 ng of DNA, 4 μL of BigDye Terminator v1.1 cycle sequencing kit (Life Technologies, Grand Island, NY, USA), and 1 μM of forward primer. Reverse reactions were also performed using reverse nested primers for each nested PCR product. Components were denatured at 96°C for 30 seconds, annealed at 50°C for 15 seconds, and extended at 60°C for 4 minutes, for a total of 25 cycles. Samples were run through Centri-Sep columns (Princeton Separations, Adelphia, NJ, USA) to remove unincorporated nucleotides and then lyophilized. Purified DNA samples were prepared for sequencing and were electrophoresed as previously described. Using Sequencing Analysis version 5.3.1 (Applied Biosystems) and Geneious v4.6.4 (Biomatters, Auckland, New Zealand) software, forward and reverse DNA electropherograms were aligned and edited. Consensus sequence was then compared with normal BRCA2 sequence (GenBank accession # NC_000013). A genogram was constructed using Geneious v4.6.4 to illustrate the inheritance of the BRCA2-3036delACAA mutation within the extended family.
Allele-specific PCR
The DNA from the woman of Scottish/Irish descent (II-8), who had breast and lung cancers, was screened for the BRCA1-c.2681-2682delAA (2800delAA) and BRCA2-c.6275_6276delTT (6503delTT) mutations. Allele-specific (AS) primers were designed to include the deletions in the forward primer for each BRCA2 site. AS primer names and nucleotide sequences are listed in Table 1. Forward primers that were normal and containing the mutation were paired with a reverse primer that matched each of their melting temperatures.
  | Table 1 Allele-specific primers used to screen two BRCA mutations seen in Great Britain populations |
The protocol for each AS PCR was the same as previously described, with one modification: annealing temperatures for BRCA1-2800delAA and BRCA2-6503delTT were 51°C and 58°C, respectively (Table 1). A positive control (human DNA containing mutation) and negative control (Nanopure water; Barnstead, Thermo Scientific) were used with each primer pair. To verify PCR results, amplicons were sequenced using the methods described earlier.
Results
BRCA2-c.2808_2811delACAA (3036delACAA) screening
Using the program Sequencing Analysis 5.3.1, each participant’s DNA sequences of the BRCA2-3036delACAA region were analyzed. As seen in Figure 2, the electropherogram on the bottom is indicative of a normal sequence that does not contain the BRCA2-3036delACAA mutation. There is a single broad peak for each base with minimal background noise. The electropherogram on top is indicative of a sequence in which a person contains one normal allele and one allele with the deletion of the four nucleotides, ACAA. The image appears to have two superimposed sequence variants. The smaller peaks beneath each larger peak represent the sequence with the deletion. It is evident that each smaller peak corresponds to the same base as the larger peak that is four bases downstream. This electropherogram pattern was observed in three family members: II-5, III-4, and III-9. The electropherograms from the rest of the participants did not have the 4-bp deletion and were considered wild-type (Wt). Reverse sequence comparisons verified each of these findings.
AS PCR screening
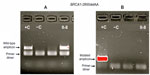
Based on AS results shown in Figure 3A and B, individual II-8 is homozygous Wt for area BRCA1-c.2681-2682delAA (2800delAA). Verification of her genotype was seen in the DNA sequence of this region. II-8’s electropherogram in Figure 4 did not have a two-base deletion and contained two adenines at the variant site. In contrast, gel images seen in Figure 5A and B illustrate that II-8 is a heterozygote for the BRCA2 gene and a carrier of the 6503delTT variant. The TT deletion was verified in the DNA sequence of this BRCA2 region, seen in Figure 6.
  | Figure 6 Electropherogram of II-8’s DNA sequence containing the BRCA2-6503delTT mutation that is aligned with the DNA sequence of a normal control. |
CHEK2 screening
DNA samples from participants were screened for founder mutations CHEK2-p.I157T and CHEK2-p.1100delC. All of the DNA sequences exhibited the Wt allele for both sites, whereas no missense mutation nor two-base deletion was observed in the electropherograms, respectively.
Discussion
The objectives of this study were to analyze three genes and mutations associated with increased risk of cancer within an extended family with a history of breast, prostate, stomach, and lung cancer. Here we report for the first time the occurrence of two different BRCA2 frameshift mutations within the same family. DNA sequence analyses of the BRCA2 gene showed that sequences of individuals who lacked the BRCA2- c.2808_2811delACAA (3036delACAA) deletion possessed only one electropherogram image, indicating two Wt alleles for those individuals. However, three individuals in this study (II-5, III-4, and III-9 from Figure 1) were carriers of the BRCA2-3036delACAA variant. The electropherograms for these individuals contained superimposed images, with one shifted by four bases downstream of the deletion site. This BRCA2 variant is one of the most common germ line mutations reported and is highly prevalent in Spanish families with histories of breast cancer.18 The deletion site is located within a hairpin secondary structure, rendering it a hotspot for mutation. In order for this four-base deletion to occur, it was predicted to have resulted from three separate mutational events.18
The BRCA2-3036delACAA mutation was found in siblings III-4 and III-9 and their paternal aunt (II-5) of Italian descent. More than likely, siblings III-4 and III-9 inherited this germ line mutation from their father, II-3, and this mutation may have contributed to the cause of their father’s prostate cancer, the father’s sister’s (II-5) two primary incidences of breast cancer, and the father’s daughter’s (III-9) two primary incidences of breast cancer. The male sibling III-4, who is 62 years of age, is at a substantially higher risk of prostate cancer. Estimates show up to one-third of male BRCA2 mutation carriers will get prostate cancer by age 65.
Participants married into the Italian bloodline do not carry this BRCA2 mutation. Since it is a rare allele in the general population, it may be safe to assume that individuals in generation IV and V have not inherited the BRCA2-3036delACAA mutation and will not be passing this allele down to any future generation.
Although the siblings of II-1 and II-2 were not screened, having breast cancer at age 46, individual III-2 may have inherited the BRCA2-3036delACAA mutation from her father (II-1). Although her father died of a brain aneurysm before age 50, he may have been a carrier of the mutation. Since individual I-1 died of stomach (and possibly pancreatic – records lack detail) cancer in his 40s, it is possible that he too was a carrier of the mutation, which may have contributed to his cancer and been passed down to at least one or perhaps more of his children. It is recommended that siblings of individual II-1 seek genetic counseling and be screened for the BRCA2-3036delACAA variant since they also have children (not illustrated in pedigree) who could have inherited this cancer-susceptibility gene.
Individual II-8, who had breast and lung cancer, was tested positive for the 6503delTT mutation by AS PCR and verified by DNA sequence comparisons. This truncated mutation most likely increased her susceptibility to breast cancer but may not have contributed to her lung cancer. Since it was revealed in her survey that she was a tobacco smoker in her earlier years, we cannot discount the possibility that two somatic mutations occurred within some other cancer-susceptibility gene. Nonetheless, it is recommended that her family members be screened for this mutation to assess their risk of breast cancer and other associated cancers. In addition, BRCA2-6503delTT screening of a tumor biopsy from II-8’s lung tissue would provide more insight into the possible association of this BRCA2 variant with lung cancer. Moreover, other founder mutations of the CHEK2 gene should be investigated as well. Finally, the BRCA2 promoter should also be screened for any possible mutation that could cause hypermethylation to occur and ultimately downregulate the BRCA2 gene, as seen in cases of non-small cell lung cancer.
In conclusion, two truncated deleterious BRCA2 mutations have been identified in a family of Italian and Irish ethnicities, with known cases of breast, prostate, stomach, and lung cancer. Germ line mutations of the BRCA2 gene substantially increase the lifetime risk of developing breast, ovarian, and prostate cancer. Since any sibling of a germ line mutation carrier has a 50% chance of inheriting the mutation, genetic testing is recommended, whereas results can be used to dictate the level and frequency of screening as well as influence decision-making on preventative measures, such as prophylactic surgery. Further studies on the possible role of BRCA2 in lung cancer need to be explored.
Acknowledgments
This project was partly funded by the University of Wisconsin-Stevens Point College of Letters and Science Summer Undergraduate Education Initiative provided by D Caporale and the UWSP Student Research Fund. DNA sequencing services were provided by the UWSP Biology Department’s Genetic Analysis Service.
Disclosure
The authors report no conflicts of interest in this work.
References
Cancer prevention and control: breast cancer [webpage on the Internet]. Atlanta, GA: Centers for Disease Control and Prevention; 2012. Available from: http://www.cdc.gov/cancer/index.htm. Accessed August 15, 2013. | |
Lim MJ, Foster GJ, Gite S, Ostendorff HP, Narod S, Rothschild KJ. An ELISA-based high throughput protein truncation test for inherited breast cancer. Breast Cancer Res. 2010;12(5):R78. | |
Chenevix-Trench G, Healey S, Lakhani S, et al; kConFab Investigators. Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res. 2006;66(4):2019–2027. | |
Lewis R. The genetics of cancer. In: Lewis R, editor. Human Genetics: Concepts and Applications. 8th ed. New York: McGraw-Hill; 2008:355–376. | |
Boettger MB, Sergi C, Meyer P. BRCA1/2 mutation screening and LOH analysis of lung adenocarcinoma tissue in a multiple-cancer patient with a strong family history of breast cancer. J Carcinog. 2003;2(1):5. | |
Marsit CJ, Liu M, Nelson HH, Posner M, Suzuki M, Kelsey KT. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: implications for treatment and survival. Oncogene. 2004;23(4):1000–1004. | |
Taron M, Rosell R, Felip E, et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet. 2004;13(20):2443–2449. | |
Lee MN, Tseng RC, Hsu HS, et al. Epigenetic inactivation of the chromosomal stability control genes BRCA1, BRCA2, and XRCC5 in non-small cell lung cancer. Clin Cancer Res. 2007;13(3):832–838. | |
Rosell R, Perez-Roca L, Sanchez JJ, et al. Customized treatment in non-small-cell lung cancer based on EGFR mutations and BRCA1 mRNA expression. PLoS One. 2009;4(5):e5133. | |
Meindl A, Ditsch N, Kast K, Rhiem K, Schmutzler RK. Hereditary breast and ovarian cancer: new genes, new treatments, new concepts. Dtsch Arztebl Int. 2011;108(19):323–330. | |
Rebbeck TR. Inherited genetic predisposition in breast cancer. A population-based perspective. Cancer. 1999;86(Suppl 11):2493–2501. | |
Purnomosari D, Paramita DK, Aryandono T, Pals G, van Diest PJ. A novel BRCA2 mutation in an Indonesian family found with a new, rapid, and sensitive mutation detection method based on pooled denaturing gradient gel electrophoresis and targeted sequencing. J Clin Pathol. 2005;58(5):493–499. | |
Cybulski C, Górski B, Huzarski T, et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet. 2004;75(6):1131–1135. | |
Vahteristo P, Bartkova J, Eerola H, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet. 2002;71(2):432–438. | |
Szabo CI, Kin MC. Population genetics of BRCA1 and BRCA2. Am J Hum Genet. 1997;60(5):1013–1020. | |
Liede A, Cohen B, Black DM, et al. Evidence of a founder BRCA1 mutation in Scotland. Br J Cancer. 2000;82(3):705–711. | |
Scottish/Northern Irish BRCA1/BRCA2 Consortium. BRCA1 and BRCA2 mutations in Scotland and Northern Ireland. Br J Cancer. 2003;88(8):1256–1262. | |
Infante M, Durán M, Acedo A, et al. The highly prevalent BRCA2 mutation c.2808_2811del (3036delACAA) is located in a mutational hotspot and has multiple origins. Carcinogenesis. 2013;34(11):2505–2511. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.