Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 8
The frequency of CYP2C19 genetic polymorphisms in Russian patients with peptic ulcers treated with proton pump inhibitors
Authors Sychev D, Denisenko N , Sizova Z, Grachev A, Velikolug K
Received 9 December 2014
Accepted for publication 17 February 2015
Published 27 May 2015 Volume 2015:8 Pages 111—114
DOI https://doi.org/10.2147/PGPM.S78986
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Martin Bluth
DA Sychev,1,2 NP Denisenko,1,2 ZM Sizova,2 AV Grachev,3 KA Velikolug4
1Russian Medical Academy of Post-Graduate Education, 2I.M. Sechenov First Moscow State Medical University, 3SM-Clinic, 4Out-patient department Number 51 branch 3, Moscow, Russia
Introduction: Proton pump inhibitors, which are widely used as acid-inhibitory agents for the treatment of peptic ulcers, are mainly metabolized by 2C19 isoenzyme of cytochrome P450 (CYP2C19). CYP2C19 has genetic polymorphisms, associated with extensive, poor, intermediate or ultra-rapid metabolism of proton pump inhibitors. Genetic polymorphisms of CYP2C19 could be of clinical concern in the treatment of peptic ulcers with proton pump inhibitors.
Aim: To investigate the frequencies of CYP2C19*2, CYP2C19*3, and CYP2C19*17 alleles and genotypes in Russian patients with peptic ulcers.
Methods: We retrospectively reviewed the cases of 971 patients of Caucasian origin with Russian nationality from Moscow region with endoscopically and histologically proven ulcers, 428 males (44%) and 543 females (56%). The mean age was 44.6±11.9 years (range: 15–88 years). DNA was extracted from ethylenediaminetetraacetic acid whole blood samples (10 mL). The polymorphisms CYP2C19 681G>A (CYP2C19*2, rs4244285), CYP2C19 636 G>A (CYP2C19*3, rs4986893) and CYP2C19 -806 C>T (CYP2C19*17, rs12248560) were evaluated using real-time polymerase chain reaction.
Results: Regarding CYP2C19 genotype, 317 patients (32.65%) out of 971 were CYP2C19*1/*1 carriers classified as extensive metabolizers. Three hundred and eighty-six (39.75%) with CYP2C19*1/*17 or CYP2C19*17/*17 genotype were ultra-rapid metabolizers. Two hundred and fifty-one people (25.85%) were intermediate metabolizers with CYP2C19*1/*2, CYP2C19*2/*17, CYP2C19*1/*3, CYP2C19*3/*17 genotypes. Seventeen patients (1.75%) with CYP2C19*2/*2, CYP2C19*3/*3, CYP2C19*2/*3 genotypes were poor metabolizers. The allele frequencies were the following: CYP2C19*2 – 0.140, CYP2C19*3 – 0.006, CYP2C19*17 – 0.274.
Conclusion: There is a high frequency of CYP2C19 genotypes associated with modified response to proton pump inhibitors in Russian patients with peptic ulcers. Genotyping for CYP2C19 polymorphisms is suggested to be a useful tool for personalized dosing of proton pump inhibitors.
Keywords: CYP2C19, proton pump inhibitors, peptic ulcer, Russian
Introduction
Cytochrome P450 is known to be a major enzyme responsible for the oxidative metabolism of drugs in the liver. Proton pump inhibitors, which are widely used as acid-inhibitory agents for the treatment of peptic ulcers, are mainly metabolized by CYP2C19, and by CYP3A4 to a lesser extent. CYP2C19 is the main isoenzyme which is related to the metabolism of omeprazole and esomeprazole, while both CYP2C19 and CYP3A4 undertake the metabolism of pantoprazole and lansoprazole. As for rabeprazole it undergoes non-enzymatic conversion and therefore has less reliance on CYP2C19. All the metabolites produced are inactive.1,2
CYP2C19 has genetic polymorphisms that correlate with its activity. All the individuals may be classified as extensive metabolizers (EMs), poor metabolizers (PMs), intermediate metabolizers (IMs), and ultra-rapid metabolizers (UMs) according to their CYP2C19-based ability to metabolize proton pump inhibitors.3,4
Individuals with CYP2C19*1/*1 genotype are EMs, they carry two wild-type alleles and have no mutation, whereas those who have CYP2C19*2 (681G>A) or CYP2C19*3 (636 G>A) mutation alleles are designated as IMs, if heterozygous, and PMs, if homozygous. IMs and especially PMs have superior acid suppression with conventional doses of proton pump inhibitors.5–7 The CYP2C19*17 (−806 C>T) allele, a novel allele identified by Sim et al8 is associated with ultra-rapid phenotype and provides decreased acid suppression with standard recommended doses of proton pump inhibitors.9–11 The areas under the concentration-time curve of proton pump inhibitors are higher in PMs than in EMs, whereas IMs are intermediate between the two; UMs have the lowest area under the concentration-time curve. Thus, proton pump inhibitors may not provide sufficient acid suppression in EMs and especially UMs.
Genotyping for CYP2C19 polymorphisms reveals considerable interethnic differences. Most PMs are to be found among Asian and African-American population (allele frequencies are 30% and 17% for CYP2C19*2; 5% and 0.4% for CYP2C19*3, respectively). CYP2C19*2 is also relatively common in Caucasians with an allele frequency of 15% compared to 0.04% for CYP2C19*3 allele.2 Thus, genetic polymorphism of CYP2C19 could be of clinical concern in the treatment of peptic ulcers with proton pump inhibitors.
The aim of the study – to investigate the frequencies of CYP2C19*2, CYP2C19*3, and CYP2C19*17 alleles and genotypes in Russian patients with peptic ulcers.
Methods
We retrospectively reviewed the cases of 971 patients of Caucasian origin with Russian nationality from the Moscow region with endoscopically and histologically proven ulcers, 428 males (44%) and 543 females (56%). The mean age was 44.6±11.9 years (range: 15–88 years). We selected patients who were tested for CYP2C19 polymorphism. In all patients, CYP2C19 polymorphism testing and gastroduodenoscopy with biopsy were done after informed consent was obtained.
DNA was extracted from ethylenediaminetetraacetic acid whole blood samples (10 mL) and isolated from peripheral lymphocytes. The polymorphisms CYP2C19 681G>A (CYP2C19*2, rs4244285), CYP2C19 636 G>A (CYP2C19*3, rs4986893), and CYP2C19 −806 C>T (CYP2C19*17, rs12248560) were evaluated using real-time polymerase chain reaction. We used LightMix for CYP2C19*2 and CYP2C19*3 and LightSNiP for CYP2C19*17 (Roche®; Hoffman-La Roche Ltd, Basel, Switzerland) in a LightCycler 2.0 system.12
Patients were referred to as EMs, PMs, IMs or UMs according to the Dutch Pharmacogenetics Working Group Guideline of the Royal Dutch Pharmacists Association (http://www.pharmgkb.org/guideline/PA166104957).
The correspondence of the distribution of the genotype frequencies to the Hardy-Weinberg equilibrium was assessed using a χ2 test with Yates correction. A P-value <0.05 was considered statistically significant. The statistical analysis was performed using SPSS Statistic 20.
Results
Regarding CYP2C19 genotype, 317 patients out of 971 were CYP2C19*1/*1 carriers classified as EMs, 386 with CYP2C19*1/*17 or CYP2C19*17/*17 genotype were UMs, 251 were IMs, and 17 were PMs (Table 1).
  | Table 1 CYP2C19 genetic polymorphisms frequency in Russian patients with peptic ulcers |
The allele frequencies were the following: CYP2C19*2 – 0.140, CYP2C19*3 – 0.006, CYP2C19*17 – 0.274 (Table 2). Genotype frequencies are summarized in Table 3.
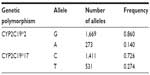  | Table 2 CYP2C19 allele frequency in Russian patients with peptic ulcers |
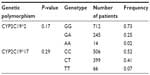  | Table 3 CYP2C19 genotype frequency in Russian patients with peptic ulcers |
The observed genotype frequencies were in the Hardy-Weinberg equilibrium (P>0.05) for the majority of CYP2C19 polymorphisms. We did not calculate CYP2C19*3 deviation from the Hardy-Weinberg equilibrium because CYP2C19*3 mainly occurs in Asian populations and is rare in Caucasians.
Discussion
The incidence of peptic ulcer in different countries of the world varies from 5% to 15%. It has been reported that CYP2C19 polymorphism affects cure rates of peptic ulcers.4–7 CYP2C19*2 and CYP2C19*3 alleles are associated with high eradication rates of Helicobacter pylori in patients with peptic ulcers, when triple therapy is administered.6 The data on CYP2C19*17 allele are very scarce. It is reported, that patients carrying ultra-rapid CYP2C19*17 allele have lower levels of proton pump inhibitors in plasma.7
Our study provides data on CYP2C19 genetic polymorphisms and is the first that has investigated CYP2C19*17 allele frequency in Russian patients with peptic ulcers.
We compared CYP2C19*2 allele and genotype frequency in our study and in Gaikovitch et al’s study on healthy volunteers from Voronezh area, Russia, and no statistically significant difference was found, P>0.05 (Tables 4 and 5).13
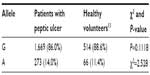  | Table 4 CYP2C19*2 allele frequency in Russian patients with peptic ulcers compared to healthy Russian volunteers |
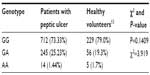  | Table 5 CYP2C19*2 genotype frequency in Russian patients with peptic ulcers compared to healthy Russian volunteers |
The frequency of CYP2C19*2 allele in our study is also comparable with data described for healthy volunteers from Slovenia (15.9%), Italy (9.4%), Croatia (15%), the Netherlands (13.3%), Germany (15%), and Greece (13.1%).14–19
CYP2C19*17 allele frequency in Russian patients tended to be higher than in Swedish (18%) and Chinese (4%) populations, thus a standard dose of proton pump inhibitors may not be effective in this group.8
Conclusion
There is a high frequency of CYP2C19 genotypes associated with modified response to proton pump inhibitors in Russian patients with peptic ulcers. Genotyping for CYP2C19 polymorphisms is suggested to be a useful tool for personalized dosing of proton pump inhibitors.
Disclosure
The authors declare no conflicts of interest.
References
Hagymási K, Müllner K, Herszényi L, Tulassay Z. Update on the pharmacogenomics of proton pump inhibitors. Pharmacogenomics. 2011;12(6):873–888. | |
Gardiner SJ, Begg EJ. Pharmacogenetics, Drug-Metabolizing Enzymes, and Clinical Practice. Pharmacol Rev. 2006;58(3):521–590. | |
Furuta T, Shirai N, Sugimoto M, Ohashi K, Ishizaki T. Pharmacogenomics of table proton pump inhibitors. Pharmacogenomics. 2004;5(2):181–202. | |
Chaudhry AS, Kochhar R, Kohli KK. Genetic polymorphism of CYP2C19 and therapeutic response to proton pump inhibitors. Indian J Med Res. 2008;127(6):521–530. | |
Furuta T, Shirai N, Takashima M, et al. Effects of genotypic differences in CYP2C19 status on cure rates for Helicobacter pylori infection by dual therapy with rabeprazole plus amoxicillin. Pharmacogenetics. 2001;11(4):341–348. | |
Furuta T, Shirai N, Takashima M, et al. Effect of genotypic differences in CYP2C19 on cure rates for Helicobacter pylori infection by triple therapy with a proton pump inhibitor, amoxicillin, and clarithromycin. Clin Pharmacol Ther. 2001;69(3):158–168. | |
Gawrońska-Szklarz B, Siuda A, Kurzawski M, Bielicki D, Marlicz W, Droździk M. Effects of CYP2C19, MDR1, and interleukin 1-B gene variants on the eradication rate of Helicobacter pylori infection by triple therapy with pantoprazole, amoxicillin, and metronidazole. Eur J Clin Pharmacol. 2010;66(7):681–687. | |
Sim S, Risinger C, Dahl ML, et al. Common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79(1):103–113. | |
Yang JC, Lin CJ. CYP2C19 genotypes in the pharmacokinetics/pharmacodynamics of proton pump inhibitor-based therapy of Helicobacter pylori infection. Expert Opin Drug Metab Toxicol. 2010;6(1):29–41. | |
Baldwin RM, Ohlsson S, Pedersen RS, et al. Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers. Br J Clin Pharmacol. 2008;65(5):767–774. | |
Furuta T, Sugimoto M, Shirai N. Individualized therapy for gastroesophageal reflux disease: potential impact of pharmacogenetic testing based on CYP2C19. Mol Diagn Ther. 2012;16(4):223–234. | |
De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46(4):594–598. | |
Gaikovitch EA, Cascorbi I, Mrozikiewicz PM, et al. Polymorphisms of drug-metabolizing enzymes CYP2C9, CYP2C19, CYP2D6, CYP1A1, NAT2 and of P-glycoprotein in a Russian population. Eur J Clin Pharmacol. 2003;59(4):303–312. | |
Herman D, Dolzan V, Breskvar K. Genetic polymorphism of cytochromes P450 2C9 and 2C19 in Slovenian population. Zdrav Vestn. 2003;72:347–351. | |
Scordo MG, Caputi AP, D’Arrigo C, Fava G, Spina E. Allele and genotype frequencies of CYP2C9, CYP2C19 and CYP2D6 in an Italian population. Pharmacol Res. 2004;50(2):195–200. | |
Arvanitidis K, Ragia G, Iordanidou M, et al. Genetic polymorphisms of drug-metabolizing enzymes CYP2D6, CYP2C9, CYP2C19 and CYP3A5 in the Greek population. Fundam Clin Pharmacol. 2007;21(4):419–426. | |
Bozina N, Granić P, Lalić Z, Tramisak I, Lovrić M, Stavljenić-Rukavina A. Genetic polymorphisms of cytochromes P450: CYP2C9, CYP2C19, and CYP2D6 in Croatian population. Croat Med J. 2003;44(4):425–428. | |
Tamminga WJ, Wemer J, Oosterhuis B, de Zeeuw RA, de Leij LF, Jonkman JH. The prevalence of CYP2D6 and CYP2C19 genotypes in a population of healthy Dutch volunteers. Eur J ClinPharmacol. 2001;57(10):717–722. | |
Brockmöller J, Rost KL, Gross D, Schenkel A, Roots I. Phenotyping of CYP2C19 with enantiospecific HPLC-quantification of R- and S-mephenytoin and comparison with the intron4/exon5 G-->A-splice site mutation. Pharmacogenetics. 1995;5(2):80–88. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
