Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Self-assembled nanoparticles based on the c(RGDfk) peptide for the delivery of siRNA targeting the VEGFR2 gene for tumor therapy
Authors Liu L, Liu X, Xu Q, Wu P, Zuo X, Zhang J, Deng H, Wu Z, Ji A
Received 7 March 2014
Accepted for publication 3 May 2014
Published 29 July 2014 Volume 2014:9(1) Pages 3509—3526
DOI https://doi.org/10.2147/IJN.S63717
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Li Liu,1 Xiaoxia Liu,1 Qian Xu,1 Ping Wu,2 Xialin Zuo,3 Jingjing Zhang,1 Houliang Deng,1 Zhuomin Wu,1 Aimin Ji1
1Department of Pharmacy, Zhujiang Hospital, Southern Medical University, Guangzhou, People’s Republic of China; 2Department of Pharmacy, Chengdu Integrated TCM & Western Medicine Hospital, Chengdu, People’s Republic of China; 3Institute of Neurosciences and the Second Affiliated Hospital of Guangzhou Medical University, Key Laboratory of Neurogenetics and Channelopathies of Guangdong Province and the Ministry of Education of China, Guangzhou, People’s Republic of China
Abstract: The clinical application of small interfering RNA (siRNA) has been restricted by their poor intracellular uptake, low serum stability, and inability to target specific cells. During the last several decades, a great deal of effort has been devoted to exploring materials for siRNA delivery. In this study, biodegradable, tumor-targeted, self-assembled peptide nanoparticles consisting of cyclo(Arg–Gly–Asp–d–Phe–Lys)-8–amino–3,6–dioxaoctanoic acid–β–maleimidopropionic acid (hereafter referred to as RPM) were found to be an effective siRNA carrier both in vitro and in vivo. The nanoparticles were characterized based on transmission electron microscopy, circular dichroism spectra, and dynamic light scattering. In vitro analyses showed that the RPM/VEGFR2-siRNA exhibited negligible cytotoxicity and induced effective gene silencing. Delivery of the RPM/VEGFR2 (zebrafish)-siRNA into zebrafish embryos resulted in inhibition of neovascularization. Administration of RPM/VEGFR2 (mouse)-siRNA to tumor-bearing nude mice led to a significant inhibition of tumor growth, a marked reduction of vessels, and a downregulation of VEGFR2 (messenger RNA and protein) in tumor tissue. Furthermore, the levels of IFN-α, IFN-γ, IL-12, and IL-6 in mouse serum, assayed via enzyme-linked immunosorbent assay, did not indicate any immunogenicity of the RPM/VEGFR2 (mouse)-siRNA in vivo. In conclusion, RPM may provide a safe and effective delivery vector for the clinical application of siRNAs in tumor therapy.
Keywords: siRNA delivery, self-assembly nanoparticles, gene silencing, tumor targeting
Introduction
Double-stranded, small interfering RNA (siRNA)-induced gene silencing through the inhibition of specific messenger RNA (mRNA) translation, also known as RNA interference, has been utilized for quite some time.1 siRNA has attracted intense interest due to its promising therapeutic effects in various diseases, such as neuronal diseases, infectious diseases, and various cancers.2,3 However, siRNA technology still faces a series of obstacles before it can be applied in a clinical setting, related to issues such as poor pharmacokinetics profiles2 due to degradation by nucleases in the serum, poor cellular uptake, rapid elimination, and the inability to target specific cell types. Therefore, designing carriers that can effectively deliver specific siRNAs to targeted tissues represents a great challenge and is the subject of intense research. Many nonviral carriers that can self-assemble into supramolecular complexes have been designed for siRNA delivery to date. For example, liposomes, lipoplexes, stable nucleic acid lipid particles, cationic polymers, and peptides have been employed to protect siRNAs from undesirable degradation during the transfection process.4 Additionally, these carriers have been modified with different targeting ligands, such as the Arg-Gly-Asp (RGD) peptide,5 folic acid,6 transferrin protein,7 and antibodies,8 to increase their targeting ability. The RGD peptide and structurally related compounds9–14 are the best-studied ligands that belong to the integrin ligand group.15–18 Because these ligands specifically bind to the integrin receptor, which is overexpressed in the endothelial cells of the tumor neovasculature,19 when applied in vivo, an 8-amino-3,6-dioxaoctanoic acid (PEG)-β-maleimidopropionic acid (MAL) hydrophilically modified, specific integrin αvβ3 receptor-targeted small cyclopeptide c(RGDfk) could lead to the accumulation of siRNA in tumors, resulting in tumor targeting.
Inhibition of angiogenesis, which blocks the supply of nutrition to and waste discharge from tumors, results in inhibition of the growth, invasion, and metastasis of tumors and has been widely applied in antitumor studies.20,21 Vascular endothelial growth factor (VEGF), also known as vascular permeability factor, plays a vital role in the angiogenic process by binding to the specific VEGF receptor 2 (VEGFR2, also known as KDR/Flk-1), a tyrosine kinase receptor, which then activates downstream signaling pathways and results in the proliferation and migration of endothelial vessels, consequently promoting angiogenesis and vascular growth.22–26 Therefore, inhibition of VEGFR2 mRNA expression in new vessels is an effective method of tumor therapy.
In the present study, RPM was found to self-assemble into nanoparticles (NPs) that could be used for efficient siRNA delivery. We examined the characteristics of the NPs and validated their function by studying the gene-silencing effects of RPM/VEGFR2-siRNA both in vitro and in vivo. We achieved two levels of targeting: targeted binding to the integrin αvβ3 receptor, which is overexpressed in new vessels, via the ligand cyclo(RGD-d-Phe-Lys) (c[RGDfk]) and gene pathway selectivity via the siRNA oligonucleotide. To our knowledge, this is the first study to show that the modified small cyclopeptide c(RGDfk) has the ability to self-assemble and can effectively deliver siRNA to targeted tissue sites.
Materials and methods
Materials
c(RGDfk)-PEG-MAL and cyclo(Arg-Ala-Asp-d-Phe-Lys) (c[RADfk])-PEG-MAL (non-targeted control peptide, hereafter referred to as RAPM) were purchased from Peptides International, Inc. (Louisville, KY, USA). Lipofectamine™ 2000 (Lipo2000), Opti-MEM®, Dulbecco’s modified Eagle’s Medium (DMEM), fetal bovine serum (FBS), and an antibiotic-antimycotic solution were purchased from Thermo Fisher Scientific (Waltham, MA, USA). The following siRNA sequences were used in the in vitro experiments: anti-human VEGFR2 siRNA (sense strand, 5′-GGUAAAGAUUGAUGAAGAAdTdT-3′, and antisense strand, 3′-dTdTCCAUUUCUAACUACUUCUU-5′); and scramble siRNA, referred to as control siRNA (sense strand, 5′-CCUGGAGAAUCAGACGACAAGUAUU-3′, and antisense strand, 3′-GGACCUCUUAGUCUGCUGUUCAUAA-5′). The following siRNA sequences were employed in the in vivo experiments: anti-mouse VEGFR2 siRNA, which was 2′-o-methyl sugar modified (sense strand, 5′-CGGAGAAGAAUGUGGUUAAdTdT-3′, and antisense strand, 3′-dTdTGCCUCUUCUUACACCAAUU-5′); anti-zebrafish VEGFR2 siRNA (sense strand, 5′-CUGAAAACAAUGUUGUGAAdTdT-3′, and antisense strand, 3′-dTdTGACUUUUGUUACAACACUU-5′); and control siRNA (mouse, zebrafish) (sense strand, 5′-CGUGAUUGCGAGACUCUGAdTdT-3′, and antisense strand, 3′-dTdTGCACUAACGCUCUGAGACU-5′). Indodicarbocyanine-5 (Cy5)-labeled siRNA (siRNA-Cy5) and all of the abovementioned siRNAs were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, People’s Republic of China). The siRNA-Cy5 was synthesized on the solid support using Cy5-phosphoramidite. Standard coupling conditions for synthesis of Cy5 labeling was carried out at the 5′-end of the guide (antisense) strand of the molecules, which formed a phosphodiester linkage. The siRNA-Cy5 was only used for the targeting research in vitro and biodistribution in vivo.
Synthesis of RPM and RAPM
RPM (96.8% purity) and RAPM (96.9%) were produced and purified by Peptides International, Inc. RPM consisted of c(RGDfk) peptide, PEG, and MAL, while RAPM consisted of c(RADfk) peptide, PEG, and MAL.
Preparation of RPM NPs and RPM/siRNA complexes
Lyophilized RPM powder was dissolved in double-distilled water (ddH2O) at a concentration of 2 mg/mL, and the siRNAs were dissolved in RNAse-free ddH2O at a concentration of 20 μM. For siRNA loading, RPM was added to the siRNA solution in different proportions (Table 1) to obtain the optimal mixture. After gentle mixing, the solution was incubated at room temperature for 30 minutes prior to the next steps.
Characterizations of RPM NPs and complexes
The particle size and zeta potential of the RPM NPs and RPM/siRNA complexes were both measured by dynamic light scattering (DLS) using a Malvern Zetasizer (Nano-ZS90; Malvern Instruments Ltd., Malvern, UK). Samples were prepared as described in the Preparation of RPM NPs and RPM/siRNA complexes section. The volume ratio of RPM (2 mg/mL, 750 μL) and siRNA (20 μM, 50 μL) was 1.5:1; the detection volume of both solutions was 1 mL; and the final concentration of siRNA in RPM/siRNA group was 100 nM. The morphology of the NPs and the NP complexes were observed using a transmission electron microscope ([TEM] JEM-100CXII; JEOL, Tokyo, Japan). About 20 μL of each sample was placed on the carbon-coated copper grids respectively, negatively stained with 2% (w/v) phosphotungstic acid, and then dried at room temperature. The secondary structures of the complexes were detected based on circular dichroism spectra (using a JASCO J-810 spectropolarimeter; Jasco, Inc., Eastern, Md, Japan) and scanned at 0.1 nm intervals from 250 to 190 nm (0.1 mm quartz cuvette). The detection volume was 1 mL for both groups. The ellipticity measurements are presented as the mean residue ellipticity ([θ] in degrees cm2 dmol-1).
Gel retardation assay
NP complexes were prepared at different ratios. All the samples contained 2 μL of siRNA (20 μM) and 1 μL, 2 μL, 3 μL, and 4 μL of RPM (2 mg/mL) were used, respectively. The complexes mixed with DNA loading buffer (6×) were then electrophoresed in a 2% agarose gel containing 0.5 μg/mL ethidium bromide. Electrophoresis was performed at 80 mV for 30 minutes, and the resulting gels were photographed under ultraviolet illumination. Naked siRNA was used as a control.
Serum stability analysis
Equivalent samples of naked siRNA (ddH2O: 3 μL; siRNA: 2 μL, 20 μm) or RPM/siRNA complexes (RPM: 3 μL, 2 mg/mL; siRNA: 2 μL, 20 μM) were mixed in a 1:1 volume ratio with FBS to achieve a 50% serum concentration and then incubated at 37°C. The final volume for all the samples was 10 μL. Samples were collected at different time points, and electrophoresed in a 2% agarose gel.
Cell culture
Human umbilical vein endothelial cells (HUVECs) (American Type Culture Collection [ATCC], Manassas, VA, USA) were cultured in DMEM supplemented with 10% FBS and a 1% antibiotic-antimycotic solution. The human non-small-cell lung cancer A549 cell line (ATCC) and the luciferase-A549 cell line (a generous gift from the Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Science) were cultured in Roswell Park Memorial Institute-1640 medium supplemented with 10% FBS and a 1% antibiotic-antimycotic solution. The cells were incubated at 37°C in a 5% CO2 atmosphere. The cells for all experiments were in logarithmic growth phase.
Confocal laser scanning
HUVECs (8×104 cells/well) and A549 cells (8×104 cells/well) were seeded in confocal laser Petri dishes (Corning Incorporated, Corning, NY, USA) and incubated for 24 hours for the next experiments. Different RPM/siRNA-Cy5 complexes (Table 1) were prepared to add to the HUVECs to observe the cellular uptake level of siRNA-Cy5 at different ratios. To observe how RPM/siRNA-Cy5 distributed in different cells, RPM/siRNA-Cy5 (RPM: 2 mg/mL, 7.5 μL; siRNA-Cy5: 20 μM, 5 μL) was added to the HUVECs and A549 cells, followed by incubation for 6 hours. To observe how the distribution of siRNA-Cy5 changed over time, the HUVECs were incubated with RPM/siRNA-Cy5 (RPM: 2 mg/mL, 7.5 μL; siRNA-Cy5: 20 μM, 5 μL) for 1 or 6 hours. To demonstrate the effect of RPM binding to the αvβ3 receptor, the HUVECs were also incubated with rabbit anti-integrin αvβ3 antibody (10 μL, 1 mg/mL) (PL Laboratories Inc., Port Moody, BC, Canada) for 30 minutes at 37°C prior to transfection with RPM/siRNA-Cy5 (RPM: 2 mg/mL, 7.5 μL; siRNA-Cy5: 20 μM, 5 μL). To throw further light on the targeting effect of RPM. As a non-targeting control, RAPM/siRNA-Cy5 (RPM: 2 mg/mL, 7.5 μL; iRNA-Cy5: 20 μM, 5 μL) was also added to the HUVECs for 6 hours incubation. The incubation volume for all the experiments above was 1 mL, and the final concentration of siRNA-Cy5 was 100 nM. Cells were fixed using 4% paraformaldehyde (Whiga Technology Co., Ltd, Guangzhou, People’s Republic of China) at room temperature for 10 minutes, and the nuclei were labeled by 4′,6-diamidino-2-phenylindole (DAPI) ([1 μg/mL] Beyotime Institute of Biotechnology, Haimen, People’s Republic of China) for 10 minutes at 37°C. Finally, dishes were observed under a confocal laser scanning microscope imaging system (Olympus Corporation, Tokyo, Japan). Cy5 excitation =640 nm, emission =680 nm.
Fluorescence-activated cell sorting analysis
To quantitatively examine the cellular uptake level of RPM/siRNA-Cy5, fluorescence-activated cell sorting (FACS) analysis was used. HUVECs were incubated with different RPM/siRNA complexes (Table 1) for 6 hours at 37°C. The incubation volume for all the groups was 1 mL, and the final concentration of siRNA-Cy5 was 100 nM (5 μL, 20 μM). Then, the cells were washed with phosphate-buffered saline (PBS) three times to remove any extracellular siRNA-Cy5, and then trypsinized and suspended in 10% FBS/DMEM to inhibit the trypsin activity. Cells were collected and suspended in 400 μL PBS within flow tubes, and cell fluorescence was determined with the BD FACSCalibur CellSorting System (model number BD FACSCALIBUR) (BD Biosciences, San Jose, CA, USA). Approximately 2×105 cells were scanned per sample using the fluorescence channel (equipped with a 660 nm band pass filter) of the FACS system. Data were obtained and analyzed using CellQuest software (version 3.1) (BD Biosciences).
Cytotoxicity
HUVECs (5×103 cells/well) were plated in 96-well plates and incubated overnight following transfection with the RPM/siRNA complexes, Lipofectamine 2000/siRNA, RPM/control siRNA complexes, or RPM NPs. Three wells were transfected for each group. The incubation volume was 100 μL and the final concentration of siRNAs was 100 nM (0.5 μL, 20 μM). According to the optimal mixture, which was selected basing on the results of confocal laser scanning and FACS, 0.75 μL RPM (2 mg/mL) was used. And 0.15 μL Lipo2000 was used according to the manufacturer’s protocol. Following 24 hours’ incubation at 37°C, cytotoxicity was assessed using the Cell Counting Kit-8 (CCK-8) assay according to the manufacturer’s instructions (Beyotime Institute of Biotechnology). Briefly, 10 μL of CCK-8 was added in each well for 2 hours’ incubation at 37°C. Then, the optical density (OD) was measured at 450 nm using a microplate reader (model 680; Bio-Rad Laboratories Inc., Hercules, CA, USA). Cytotoxicity was calculated according to the following equation:
Cell viability rate = [(ODexperiment − ODblank)/(ODcontrol − ODblank)] × 100%. | (1) |
The assays were conducted in triplicate.
NP complex-mediated gene silencing in vitro
Intracellular siRNA transfection
HUVECs were seeded into a six-well plate and incubated for 24 hours, until the cells were 60% confluent. The samples were then transfected with RPM/siVEGFR2, RPM/control siRNA, RPM NPs alone, or siVEGFR2 alone using the Lipo2000 reagent. The final volume was 1 mL and the final concentration of siRNAs was 100 nM. Untreated cells were employed as a blank control. The cellular levels of VEGFR2 mRNA were measured after 48 hours, and VEGFR2 protein levels were measured after 72 hours through quantitative reverse transcription polymerase chain reaction (RT-qPCR) and Western blot analyses, respectively.
RT-qPCR
Total RNA was extracted from the HUVECs using the TRIzol® reagent (Thermo Fisher Scientific), according to the manufacturers’ instructions. For cDNA synthesis, 2 μg of total RNA was reverse transcribed into cDNA using the ReverTra Ace® qPCR RT Master Mix with gDNA Remover kit (Toyobo, Osaka, Japan). Real-time monitoring of the PCR amplification of the cDNA was performed using a real-time PCR detection system (LightCycler® 480; Hoffman-La Roche Ltd., Basel, Switzerland) with Real Master Mix (SYBR® Green) (Toyobo). PCR amplification was conducted through 40 cycles of denaturation at 95°C for 15 seconds, followed by annealing at 60°C for 15 seconds and extension at 72°C for 32 seconds. The data were calculated using the 2−ΔΔCt method.27 The following primer sequences were employed: VEGFR2-specific primers (sense, 5′-GCCAGTCTTCTAGGCATATCC-3′, and antisense, 5′-CTCCCCAGGTACTGCTACTT-3′); GAPDH-specific primers (sense, 5′-GAACGGGAAGCTCACTGG-3′, and antisense, 5′-GCCTGCTTCACCACCTTCT-3′). GAPDH (a housekeeping gene) was used as an internal reference.
Western blot analysis
Transfected cells were collected, and Western blot analysis was conducted as described previously.28 Briefly, total protein was extracted using a total protein extraction kit (KGP250; KeyGen Biotech. Co. Ltd., Nanjing, People’s Republic of China) according to the manufacturer’s instructions. The protein concentration was determined using the bicinchoninic acid Protein Assay Kit (KeyGEN). Fifty micrograms of total protein was used in these analyses. The level of VEGFR2 protein was normalized to the level of GAPDH protein. The density of the obtained bands was scanned and calculated using Image J 1.42q software (National Institutes of Health, Bethesda, MD, USA).
In vivo angiogenesis assay in zebrafish
Flk-1-GFP transgenic and wild-type zebrafish were provided by the Department of Cell Biology of Southern Medical University, Guangzhou, People’s Republic of China. The zebrafish were raised according to the methods of Westerfield.29 Zebrafish embryos were obtained from adult mating pairs and reared in Holtfreter’s solution (60 mmol/L NaCl, 2.4 mmol/L sodium bicarbonate, 0.8 mmol/L CaCl2, 0.67 mmol/L KCl, and 10 mmol/L 4-[2-hydroxyethyl]-1-piperazineethanesulfonic acid [HEPES]) in a humidified incubator at 28.5°C. Healthy embryos were selected at 10 hours post-fertilization under a dissecting microscope, and 20 embryos/well were distributed into six-well plates. Then, RPM/siVEGFR2 (RPM: 2 mg/mL, 3 μL; siVEGFR2: 20 μM, 2 μL), RPM/control siRNA (RPM: 2 mg/mL, 3 μL; control siRNA: 20 μM, 2 μL), naked siVEGFR2 (ddH2O: 3 μL; siVEGFR2: 20 μM, 2 μL), or ddH2O was delivered to the zebrafish embryos through microinjection (2 nL per embryo). The final volume was 5 μL for all the samples. Blood vessel formation and associated fluorescence intensity were examined about 72 hours post-fertilization via confocal microscopy. Confocal laser microscopy images were processed to gray images using Adobe Photoshop software (v 7.0; Adobe Systems Incorporated, San Jose, CA, USA). Then the gray images were analyzed using Image J software. The zebrafish were anesthetized using 0.02% tricaine mesylate prior to observation.
In vivo analyses in tumor-bearing nude mice
Human non-small-cell lung cancer xenograft tumor model
This project was approved by the Zhongshan School of Medicine of Sun Yat-sen University (Guangzhou, People’s Republic of China). Xenograft tumor models of human non-small-cell lung cancer were established. BALB/c nude mice (female, 4–6 weeks old, ~20 g) were purchased from the Experimental Animal Center of Sun Yat-Sen University and maintained in a sterile environment according to standardized animal care guidelines. The experiments were carried out according to national regulations and were approved by the Southern Medical University animal experiments ethical committee. Luciferase-A549 cells (1×106) were subcutaneously injected into the right shoulder of the mice. When the tumor volume reached 40–50 mm3, the animals were randomly divided into different groups for testing. Saline, RPM/control siRNA, and RPM/siVEGFR2 were administered via intravenous injection in 100 μL volumes (RPM: 60 μL, 5 mg/mL; siRNAs: 40 μL, 50 μM) once every 3 days at a dose of 2 nmol of siRNAs per mouse, and the treatment was repeated six times. Tumor luminescence was detected on day 0 and day 20 to monitor the development of the tumors using the In Vivo Imaging Systems Spectrum imaging system (Xenogen Corporation–Caliper, Ala-meda, CA, USA) following intraperitoneal injection of luciferin (Promega Corporation, Madison, WI, USA) at a dose of 150 mg/kg per mouse. Tumor volume and mouse weight were monitored prior to injection. The tumor volume was calculated using the following formula:
Volume = ½ × length × width2. | (2) |
The curves were plotted as the mean tumor volume ± standard deviation. The animals were euthanized 2 days after the last treatment, and the tumors were excised and preserved in liquid nitrogen.
In vivo localization of NP complexes with siRNA-Cy5
For the tissue distribution analysis, the successful model animals were randomly divided into three groups (N=3). A single dose of saline, RPM/siRNA-Cy5 (0.2 nmol), RAPM/siRNA-Cy5 (0.2 nmol), or naked siRNA-Cy5 (0.2 nmol) was injected via the tail vein in 40 μL volumes (RPM: 30 μL, 2 mg/mL; siRNA-Cy5: 10 μL, 20 μM). The subsequent biodistribution was monitored at 10 minutes, 0.5 hours, 3 hours, 6 hours, and 24 hours after administration using the IVIS Spectrum imaging system at the appropriate wavelength (siRNA-Cy5: λex =640 nm, λem =680 nm). The mice were sacrificed after the last time point, and the tumors and major organs were excised and imaged. The uptake level of siRNA-Cy5 in different organs was quantified by doing region of interest.
Detection of VEGFR2 mRNA and protein expression in tumor tissue
Total RNA was extracted from the tumor tissues, and RT-qPCR was performed using the same protocol described above. The following primer pairs were used in this experiment: VEGFR2 sense, 5′-AGAATGCGGGCTCCTGACTA-3′, and antisense, 5′-CCATGCTCAGTGTCTCTGACA-3′; and 18s sense, 5′-CCTGGATACCGCAGCTAGGA-3′, and antisense, 5′-GCGGCGCAATACGAATGCCCC-3′. The expression of the 18s gene was used as an internal reference. The VEGFR2 protein expression level was detected through Western blot analysis.
Immunohistochemical analysis
The tumors were frozen and cut into 6 μM sections after being embedded in Tissue-Tek® optimum cutting temperature medium (Sakura Finetek Japan, Tokyo, Japan). The sections were fixed in cold 100% acetone for 10 minutes, then stored at −20°C. Prior to analysis, the sections were removed and allowed to acclimate to room temperature. The sections were then washed with PBS and incubated in endogenous peroxidase (3% H2O2 ) for 10 minutes, followed by 1% normal goat serum in PBS for 30 minutes at room temperature. A rat anti-mouse CD31 antibody (1:50; BD Biosciences) was added, and the sections were incubated overnight at 4°C. The sections were subsequently washed three times in PBS, followed by incubation with rabbit anti-rat IgG-biotin ([1:100 dilution] Wuhan Boster Biological Technology, Ltd, People’s Republic of China) at 37°C for 1 hour, after which they were washed with PBS again and incubated with peroxidase-conjugated streptavidin ([1:100 dilution] Proteintech Group, Inc., Chicago, IL, USA). The chromogen DAB Kit (Boster) and hematoxylin were used to stain vessels and nuclei. A fluorescence microscope (DM4000B; Leica Microsystems, Wetzlar, Germany) was employed to obtain photographs.
Mouse cytokine and interferon response
At 6 and 24 hours after administration of RPM NPs, RPM/siVEGFR2, and saline, respectively, serums were collected and assayed to detect mouse IFN-α, IFN-γ, IL-12, and IL-6 using a quantitative enzyme-linked immunosorbent assay (ELISA) kit (eBioscience, Inc., San Diego, CA, USA) according to the manufacturer’s instructions. The absorbance was read using a Multiskan™ MK3 microplate reader (Thermo Fisher Scientific) at 450 nm.
Renal and liver toxicity analysis
RPM/siRNA- and saline-treated animals were sacrificed 2 days after the last injection, and serum was collected and isolated for the measurements of alanine aminotransferase (ALT) and creatinine using an Aeroset automated analyzer (Abbott Diagnostics, Berkshire, UK) according to the manufacturer’s instructions.
Statistical analysis
Statistical analysis of the data was performed through one-way analysis of variance (ANOVA) (SPSS software, version 13.0; SPSS Inc., Chicago, IL, USA). The results are expressed as the mean ± standard error, and P<0.05 was considered statistically significant. All statistical tests were two-sided.
Results
Preparation and characterizations of NPs
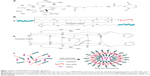
The mechanism of NP assembly is presented in Figure 1, which was speculated according to the basic characteristics that were detected by TEM, CD spectra and DLS. Through intermolecular hydrogen bonding interactions, with the β-sheet and random coil secondary structure, RPM molecules self-assembled into spherical NPs and c(RGDfk)s were on the surface of NPs for the targeting of αvβ3 receptors. siRNAs in the solution were encapsulated in the NP cavities or “trapped” in the gap of molecules due to the positively charged arginine on the c(RGDfk)s and electropositive carbonyl carbon and hydrogen atoms on the maleimide.
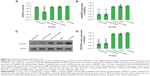
The TEM images presented in Figure 2A (inset images) show that the RPM NPs and RPM/siRNA complexes were round in shape, and their diameters were less than 100 nm. The particle size of the RPM NPs ranged from 91.28 to 122.4 nm, while the size of the RPM/siRNA complexes ranged from 105.7 to 141.8 nm (Table 2). Both types of NPs exhibited a narrow size distribution (Figure 2A) and possessed a negative charge (Table 2) (zeta potentials of −6.16 and −14.6 mV, respectively). Furthermore, Figure 2B shows a mixed secondary structure of the NPs including β-sheet and random coil, with the β-sheet accounting for the majority.
  | Table 2 The surface zeta potential of nanoparticles |
Protection of the siRNAs encapsulated in RPM
The result of gel retardation assay shows that the RPM/siRNA NPs clearly lagged behind the naked siRNA, which may be due to the disparities in their molecular weights. And with increasing the amount of RPM, more siRNAs were trapped in the NPs (Figure S1A). When the ratio reached 1.5:1, there was no difference of band intensity between naked siRNA and RPM/siRNA NPs (Figure S1B). The serum stability test demonstrated that the naked siRNA was nearly completely degraded after 4 hours of incubation in the serum. In contrast, siRNA was still present in the RPM/siRNA group after 12 hours (Figure 2C).
Intracellular distribution of siRNA
When the volume ratio reached 1.5:1, siRNA-Cy5 aggregated around the nucleus, and this phenomenon was not observed above or below this ratio (Figure 3A). Therefore, this ratio was used in the subsequent experiments as the desired ratio between RPM and the siRNAs. High-definition differential interference contrast images clearly show that siRNA-Cy5s were distributed in the cell cytoplasm (Figure 3B). The result of FACS analysis, shown in Figure S2A, was in accordance with the result of confocal microscopy images. Although no difference was found in the percentage of positive cells in different ratios (Figure S2B), when the volume ratio reached 1.5:1, the fluorescence intensity of siRNA-Cy5 in the cells was the strongest (Figure S2C).
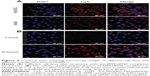
The HUVECs were incubated and maintained as indicated in the methods. As shown in Figure 4A, siRNA-Cy5s were distributed in the cytoplasm and around the nuclei in both the HUVECs and A549 cells after incubation with RPM/siRNA-Cy5 for 6 hours. In the HUVECs, the fluorescence signal from siRNA-Cy5 in the cytoplasm was weak, and puncta formed after 1 hour; however, after 6 hours of incubation, the fluorescence signal became strong and dispersed (Figure 4B), while the HUVECs treated with anti-integrin αvβ3 antibody for 30 minutes before the incubation of RPM/siRNA-Cy5 (Figure 5A), or treated with RAPM/siRNA-Cy5 (Figure 5B), showed an extremely weak fluorescent signal for siRNA-Cy5 in the cytoplasm.
Cytotoxicity
As shown in Figure 6A, there was no difference in cell viability between the RPM NP, RPM/siRNA, and untreated groups, while the cell viability observed in the Lipo2000/siRNA group was approximately 50%, which was significantly different from the other groups.
In vitro gene-silencing efficiency
RT-qPCR analysis demonstrated that transfection of siRNAs using RPM and a cationic lipid reagent resulted in comparable (~38% for RPM/siRNA versus ~39% for Lipo2000/siRNA) gene knockdown in HUVECs following treatment for 48 hours, and no significant difference was observed between other groups (Figure 6B). Total protein was collected for Western blot analysis after 72 hours, and VEGFR2 protein expression was markedly reduced in the RPM/siRNA and Lipo2000/siRNA groups (Figures 6C and 6D), by approximately 29% and 36%, respectively. Taken together, these data demonstrate the ability of the RPM/siRNA to enter the integrin αvβ3-expressing cells and induce targeted gene knockdown efficiently.
In vivo gene silencing in zebrafish
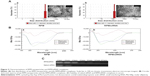
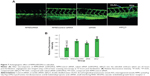
In zebrafish, hypogenetic vessels or a lack of vessels can be clearly observed because the normal vessels formed in the zebrafish express strong green fluorescent protein (GFP). As shown in Figure 7A, strong GFP fluorescence was observed in the ddH2O-treated group, while the embryos treated with the RPM/siRNA failed to completely form dorsal longitudinal anastomotic vessels and intersegmental vessels. The fluorescence statistics for each group revealed that RPM/siRNA could have an antiangiogenic effect (Figure 7B).
Tumor targeting
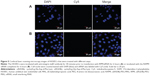
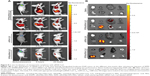
Intense fluorescence accumulated in the tumors after 0.5 hours in the RPM/siRNA-Cy5 group, reflecting the presence of siRNA-Cy5 in the blood pool, and this accumulation lasted for at least 24 hours after injection. However, no fluorescence was detected in the tumors of mice that were injected with naked siRNA-Cy5 and RAPM/siRNA-Cy5 (Figure 8A). Fluorescence was also detected in the kidneys and livers of the mice, and ex vivo images corroborated the findings of the whole-animal imaging analysis (Figure 8B). The statistic of fluorescence intensity of siRNA-Cy5 in different organs showed that siRNA-Cy5s mainly accumulated in the kidneys, then livers and tumors (Figure S3); however, there was a significant difference of fluorescence intensity in tumors between the RPM/siRNA-Cy5 group and other groups, which proved the tumor-targeting ability of RPM (**P<0.01, ##P<0.01 [**refers that there is a extreme difference between the RPM/siRNA-Cy5 group and naked siRNA-Cy5 group. ##refers that there is an extreme difference between the RPM/siRNA-Cy5 group and RAPM/siRNA-Cy5 group]).
In vivo antitumor activity
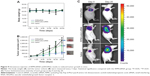
Twenty days after the first injection, no difference was observed in the body weights of the mice between the three groups (Figure 9A). An obvious tumor growth was found in the saline- and RPM/control siRNA-treated mice, and there was no significant difference between these two groups, which indicated that RPM/control siRNA did not have specific antitumor effects. Compared to the saline- and RPM/control siRNA-treated mice, the tumor volume was effectively inhibited in the mice treated with RPM/siRNA (*P<0.05, **P<0.01) (Figure 9B). Distinct attenuation of the tumor bioluminescence intensity was observed only in the RPM/siRNA group (Figure 9C).
To determine whether the anti-tumor effect of the RPM/siRNA was due to immune effects, the levels of IFN-α, IFN-γ, IL-12, and IL-6 in the serum were determined at 6 and 24 hours after administration. The results showed that there were no differences in the cytokine or interferon responses of the groups (Figure 10A). In addition, unchanged ALT and creatinine serum levels indicated that RPM/siRNA is well tolerated, producing no significant renal and liver toxicity (Figure 10B). Anti-tumor effect of RPM/siRNA was also determined at the gene level; a highly significant difference was observed in the levels of VEGFR2 mRNA between the tumors of the RPM/siRNA group and the other groups (Figure 10C). Furthermore, Figure 10D shows that the VEGFR2 protein level was markedly reduced in the RPM/siRNA group. Additionally, immunohistochemistry was performed to detect vessels in the tumors, and the density of vessels observed in the RPM/siRNA group was clearly lower than in the other two groups, as shown in Figure 10E.
Discussion
In this study, we have described a targeted type of NP and demonstrated that these NPs could effectively carry siRNA into tumors, resulting in effective gene silencing both in vitro and in vivo. First, we intuitively selected the optimal ratio of RPM and siRNA via confocal laser scanning microscopy and FACS analysis: ratios below the ideal ratio did not cause sufficient transfection effects, possibly due to a lack of RPM, while ratios above the ideal ratio were not effective due to competitive binding of the receptors by the RPM NPs and RPM/siRNA. Based on TEM and dynamic light scattering (DLS) results, a unimodal distribution was present in the RPM NP and RPM/siRNA groups. TEM showed that the NPs were spherical particles, and no distinct aggregation was observed, which may have been due to the secondary structure of the NPs. It had been proved that a helical structure can induce membrane fusion, even at physiological pH, which would be expected to induce aggregation.30,31 However, the secondary structure of the RPM NPs consisted of a β-sheet and a random coil. Because TEM was used to measure the dehydrated diameter of the NPs, while the laser particle size analyzer determined their hydrodynamic diameter, there was a slight difference in size between the results of the TEM and DLS analyses, as previously reported.32–34 The zeta potentials of the RPM NPs and RPM/siRNA were negative. In general, negatively charged particles are taken up less efficiently by cells, and they are therefore not an obvious choice for use as siRNA carriers, though they have occasionally been employed for siRNA delivery.31,35
The level of cellular uptake of NPs is always determined by three crucial factors: the zeta potential,36 particle size,37 and receptor-mediated38 internalization. Cationic NPs can effectively combine with siRNA through electrostatic attractions, whereas a small particle size is beneficial in facilitating the entry of NPs into cells through endocytosis. A targeting ligand is also required to enhance the cellular uptake of NPs, especially anionic NPs. In the present investigation, receptor-mediated internalization may have played a major role in the cellular uptake of NPs. The siRNA-Cy5s transfected into cells using RPM NPs were distributed throughout the cytoplasm and exhibited strong fluorescence. However, the fluorescence intensity of the siRNA-Cy5s in the cytoplasm was obviously reduced in cells that were initially incubated with anti-integrin αvβ3 antibody for 30 minutes; this may have been due to the competitive binding of the anti-integrin αvβ3 antibody and the RPM/siRNA-Cy5 to the integrin αvβ3 receptor. Extremely weak fluorescence in the RAPM/siRNA-Cy5 control group further demonstrated that the RPM NPs could attach to the cytomembrane and serve as an effective siRNA delivery system for integrin αvβ3-positive cells, which implies that an active transport process was used for siRNA uptake.
Due to their negative charge, our NPs did not act like cationic NPs, which often result in high cytotoxicity, especially in NPs with high zeta potentials, due to electrostatic interactions with the negatively charged glycocalyx on the cell membranes.39 An obvious cytotoxic effect of Lipo2000/siRNA was observed, which is commonly proposed to be due to the inhibition of adenosine triphosphate (ATP) enzyme activity40 and to involve the protein kinase C (PKC) pathway41 in cells. In contrast, negligible cytotoxicity was observed for our NPs, indicating the biocompatibility of the RPM NPs and demonstrating the tremendous advantages of a safe and effective siRNA delivery system. Stability of the siRNAs in serum is another indispensable condition for the clinical use of siRNAs. Due to the increased molecular weight associated with encapsulation in the RPM NPs during the self-assembly process, the RPM/siRNA complexes exhibited a lag in the gel retardation assay compared to naked siRNA. The RPM NPs protected the siRNAs from being degraded; the PEG-MAL hydrophilic modification of the NPs as well as their negative surface charge inhibits the adsorption of proteins present in serum. These results provide a theoretical basis for the in vivo application of RPM NPs in the near future.
When gene knockdown was examined in vitro, the RPM/siRNA showed a robust gene-silencing effect at both the mRNA and protein levels in HUVECs. The data also demonstrated that the RPM/siRNA displayed efficient release of the siRNAs into the cytoplasm. However, the mechanism by which the siRNAs are released from the NPs is not clear, and we speculate that dilution in the cytoplasm is the main reason for this phenomenon,42 resulting in disassociation of the RPM/siRNA and release of the siRNA. The results of the PT-qPCR and Western blot analyses indicated that off-target effects can be ignored. In summary, RPM is a useful tool for the in vitro delivery of siRNAs.
Based on the analyses described above, we next evaluated the antiangiogenic effects of RPM/siRNAs in vivo using zebrafish. It is worth noting that naked siRNA also exhibited antiangiogenic effects, similar to reports showing that injection of naked siRNA into early zebrafish embryos can lead to gene silencing through double-stranded RNA-mediated interference if the targeted gene plays a critical role during the earlier stages of development.43,44 However, a significant difference was still observed between the RPM/siRNA and naked siRNA groups (##P<0.01), demonstrating the specific, targeted gene-silencing effect of the RPM/siRNA in later developmental stages in embryos (##indicates the extremely significant difference between RPM/siRNA group and naked siRNA group).
We next evaluated the tumor-targeting effect of the RPM/siRNA in vivo. siRNA-Cy5 accumulated in tumors within 0.5 hours, and the fluorescence intensity of the siRNA-Cy5 became stronger as time elapsed. Two explanations for this phenomenon were considered. First, the delivery of the drugs to a specific site is based on the molecular recognition of an appropriate ligand.45 The c(RGDfk) peptide specifically attaches to integrin αvβ3, which is expressed on newly formed blood vessels. Thus, the RPM/siRNA might rapidly redistribute the siRNA-Cy5 from the blood pool to tumors and immobilize the NPs within the tumors.46 Second, passive targeting may also play a role in enhancing the accumulation of siRNA-Cy5 in the tumor. Previous reports have demonstrated that particles with an appropriate diameter (<200 nm) can effectively target tumors due to size-dependent properties and the enhanced permeation and retention effect of the tumor.47 Additionally, the negative charge of the NPs decreases nonspecific interactions, leading to their clearance from the circulation, thus improving the passive targeting of tumors, and the binding of the ligand and receptor might consequentially contribute to a longer retention time in the tumor’s interstitial space and an improved enhanced permeation and retention effect. Moreover, no fluorescence was detected in the tumors of mice that were injected with naked siRNA-Cy5 or RAPM/siRNA-Cy5, which further proved the tumor-targeting effect of RPM NPs. It was also noted that, 24 hours after administration of the RPM/siRNA-Cy5, the kidneys of the mice still showed strong fluorescence, indicating that the RPM/siRNA-Cy5 were undergoing renal excretion.
No reduction of body weight and renal or liver toxicity were observed in the RPM/siRNA group compared to the saline-treated group, further demonstrating that the RPM NPs exhibited no toxicity. The tumor growth curves and the downregulation of VEGFR2 mRNA and protein, as well as the decreased density of vessels, further verified the gene- and receptor-targeting effects of the RPM/siRNA. Therefore, the inhibition of vessel formation resulted in insufficient nutrition and efficiently suppressed tumor development, causing a marked reduction of the luciferase signal originating from the tumor.
Recent reports have suggested that the administration of double-stranded siRNA might induce an innate immune response, leading to nonspecific antitumor effects.48,49 Hence, IFN-α, IFN-γ, IL-12, and IL-6 were tested. The results demonstrated that neither the RPM/siRNA nor blank RPM NPs activated the innate immune response, indicating that specific gene knockdown was achieved with the RPM/siRNA.
Conclusion
We have demonstrated that RPM could self-assemble into NPs, and DLS showed that these NPs presented a narrow size distribution. TEM analyses revealed that the NPs possessed a spherical shape. In vitro assays indicated that the NPs showed good biocompatibility and negligible cytotoxicity. Due to the receptor-mediated pathway involved, the RPM/siRNA complexes exhibited effective, targeted cellular uptake and resulted in significant gene silencing in HUVECs. Moreover, no bodily toxicity or innate immune response was induced in tumor-bearing mice. Treatment with RPM/siRNA resulted in marked inhibition of tumor growth, effective gene knockdown, and antiangiogenic effects in the tumors. Collectively, RPM represents a novel delivery system for siRNAs and a promising tool for cancer therapy.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30672557), the Nature Science Foundation of Guangdong Province (8251051501000), and the Specialized Research Fund for the Doctoral Program of Higher Education (20094433110013).
Disclosure
The authors report no conflicts of interest in this work.
References
Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101(1):25–33. | ||
de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat Rev Drug Discov. 2007;6:443–453. | ||
Iorns E, Lord CJ, Turner N, Ashworth A. Utilizing RNA interference to enhance cancer drug discovery. Nat Rev Drug Discov. 2007;6: 556–568. | ||
Aliabadi HM, Landry B, Sun C, Tang T, Uludağ H. Supramolecular assemblies in functional siRNA delivery: where do we stand? Biomaterials. 2012;33:2546–2569. | ||
Danhier F, Lecouturier N, Vroman B, et al. Paclitaxel-loaded PEGylated PLGA-based nanoparticles: in vitro and in vivo evaluation. J Control Release. 2009;133:11–17. | ||
Pan J, Liu Y, Feng SS. Multifunctional nanoparticles of biodegradable copolymer blend for cancer diagnosis and treatment. Nanomedicine (Lond). 2010;5:347–360. | ||
Davis ME, Zuckerman JE, Choi CH, et al. Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nature. 2010;464:1067–1070. | ||
Alexis F, Basto P, Levy-Nissenbaum E, et al. HER-2-targeted nanoparticle-affibody bioconjugates for cancer therapy. Chem Med Chem. 2008;3: 1839–1843. | ||
Keenan RM, Callahan JF, Samanen JM, et al. Conformational preferences in a benzodiazepine series of potent nonpeptide fibrinogen receptor antagonists. J Med Chem. 1999;42(4):545–559. | ||
Fisher MJ, Gunn B, Harms CS, et al. Non-peptide RGD surrogates which mimic a Gly-Asp beta-turn: potent antagonists of platelet glycoprotein IIb-IIIa. J Med Chem. 1997;40(13):2085–2101. | ||
Zablocki JA, Miyano M, Garland RB, et al. Potent in vitro and in vivo inhibitors of platelet aggregation based upon the Arg-Gly-Asp-Phe sequence of fibrinogen. A proposal on the nature of the binding interaction between the Arg-guanidine of RGDX mimetics and the platelet GP IIb-IIIa receptor. J Med Chem. 1993;36(13):1811–1819. | ||
Egbertson MS, Chang CT, Duggan ME, et al. Non-peptide fibrinogen receptor antagonists. 2. Optimization of a tyrosine template as a mimic for Arg-Gly-Asp. J Med Chem. 1994;37(16):2537–2551. | ||
Zablocki JA, Miyano M, Garland RB, et al. Potent in vitro and in vivo inhibitors of platelet aggregation based upon the Arg-Gly-Asp sequence of fibrinogen. (Aminobenzamidino)succinyl (ABAS) series of orally active fibrinogen receptor antagonists. J Med Chem. 1995;38(13): 2378–2394. | ||
Hutchinson JH, Cook JJ, Brashear KM, et al. Non-peptide glycoprotein IIb/IIIa antagonists. 11. Design and in vivo evaluation of 3,4-dihydro-1 (1H)-isoquinolinone-based antagonists and ethyl ester prodrugs. J Med Chem. 1996;39(23):4583–4591. | ||
Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin alpha2beta1. Cell. 2000;101(1):47–56. | ||
Xiong JP, Stehle T, Zhang R, et al. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296(5565):151–155. | ||
Humphries JD, Byron A, Humphries MJ. Integrin ligands at a glance. J Cell Sci. 2006;119(19):3901–3903. | ||
Shimaoka M, Xiao T, Liu JH, et al. Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112(1):99–111. | ||
Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. | ||
Thairu N, Kiriakidis S, Dawson P, Paleolog E. Angiogenesis as a therapeutic target in arthritis in 2011: learning the lessons of the colorectal cancer experience. Angiogenesis. 2011;14(3):223–234. | ||
Hanahan D, Weinberg RA. The hall marks of cancer. Cell. 2000;100(1): 57–70. | ||
Cao R, Eriksson A, Kubo H, Alitalo K, Cao Y, Thyberg J . Comparative evaluation of FGF-2-, VEGF-A-, and VEGF-C-induced angiogenesis, lymphangiogenesis, vascular fenestrations, and permeability. Circ Res. 2004;94(5):664–670. | ||
Ferrara N. Vascular endothelial growth factor: basic science and clinical progress. Endocr Rev. 2004;25(4):581–611. | ||
Ranieri G, Patruno R, Ruggieri E, Montemurro S, Valerio P, Ribatti D. Vascular endothelial growth factor (VEGF) as a target of bevacizumab in cancer: from the biology to the clinic. Curr Med Chem. 2006;13(16): 1845–1857. | ||
Ramsden JD. Angiogenesis in the thyroid gland. J Endocrinol. 2000;166:475–480. | ||
Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. | ||
Yang J, Mani SA, Donaher JL, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117: 927–939. | ||
Chen S, Liu S, Gong W, et al. Combination therapy with VEGFR2 and EGFR siRNA enhances the antitumor effect of cisplatin in non-small cell lung cancer xenografts. Oncol Rep. 2013;29:260–268. | ||
Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of zebrafish Danio* (Brachydanio) rerio. 4th Ed. [Eugene, Or.]: ZFIN; 2000. Available from: http://zfin.org/zf_info/zfbook/zfbk.html. Accessed July 16, 2014. | ||
Subbarao NK, Parente RA, Szoka FC Jr, Nadasdi L, Pongracz K. pH-dependent bilayer destabilization by an amphipathic peptide. Biochemistry. 1987;26(11):2964–2972. | ||
Sakurai Y, Hatakeyama H, Sato Y, et al. Endosomal escape and the knockdown efficiency of liposomal-siRNA by the fusogenic peptide shGALA. Biomaterials. 2011;32:5733–5742. | ||
Zhao ZX, Gao SY, Wang JC, et al. Self-assembly nanomicelles based on cationic mPEG-PLA-b-Polyarginine(R15) triblock copolymer for siRNA delivery. Biomaterials. 2012;33:6793–6807. | ||
Gao J, Liu W, Xia Y, et al. The promotion of siRNA delivery to breast cancer overexpressing epidermal growth factor receptor through anti-EGFR antibody conjugation by immunoliposomes. Biomaterials. 2011; 32:3459–3470. | ||
Jain N, Wang Y, Jones SK, Hawkett BS, Warr GG. Optimized steric stabilization of aqueous ferrofluids and magnetic nanoparticles. Langmuir. 2010;26(6):4465–4472. | ||
Hsu SH, Ho TT, Tseng TC. Nanoparticle uptake and gene transfer efficiency for MSCs on chitosan and chitosan-hyaluronan substrates. Biomaterials. 2012;33:3639–3650. | ||
Thomas M, Klibanov AM. Non-viral gene therapy: polycation-mediated DNA delivery. Appl Microbiol Biotechnol. 2003;62:27–34. | ||
Green JJ, Chiu E, Leshchiner ES, Shi J, Langer R, Anderson DG. Electrostatic ligand coatings of nanoparticles enable ligand-specific gene delivery to human primary cells. Nano Lett. 2007;7:874–879. | ||
Arote R, Hwang SK, Lim HT, et al. The therapeutic efficiency of FP-PEA/TAM67 gene complexes via folate receptor-mediated endocytosis in a xenograft mice model. Biomaterials. 2010;31:2435–2445. | ||
Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24:1121–1131. | ||
Datiles MJ, Johnson EA, McCarty RE. Inhibition of the ATPase activity of the catalytic portion of ATP synthases by cationic amphiphiles. Biochim Biophys Acta. 2008;1777:362–368. | ||
Bottega R, Epand RM. Inhibition of protein kinase C by cationic amphiphiles. Biochemistry. 1992;31:9025–9030. | ||
Li Q, Wang J, Shahani S, et al. Biodegradable and photocrosslinkable polyphosphoester hydrogel. Biomaterials. 2006;27:1027–1034. | ||
Wargelius A, Ellingsen S, Fjose A. Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem Biophys Res Commun. 1999;263:156–161. | ||
Wang L, Zhou JY, Yao JH, Lu DR, Qiao XJ, Jia W. U6 promoter-driven siRNA injection has nonspecific effects in zebrafish. Biochem Biophys Res Commun. 2010;391:1363–1368. | ||
Byrne JD, Betancourt T, Brannon-Peppas L. Active targeting schemes for nanoparticle systems in cancer therapeutics. Adv Drug Deliv Rev. 2008;60:1615–1626. | ||
Pirollo KF, Chang EH. Does a targeting ligand influence nanoparticle tumor localization or uptake? Trends Biotechnol. 2008;26(10): 552–558. | ||
Liu P, Yu H, Sun Y, Zhu M, Duan Y. A mPEG-PLGA-b-PLL copolymer carrier for adriamycin and siRNA delivery. Biomaterials. 2012;33: 4403–4412. | ||
Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. | ||
Marques J, Williams BR. Activation of the mammalian immune system by siRNAs. Nat Biotechnol. 2005;23:1399–1405. |
Supplementary materials
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.