Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Physicochemical features and transfection properties of chitosan/poloxamer
188/poly(D,L-lactide-co-glycolide) nanoplexes
Authors Cosco D , Federico C, Maiuolo J, Bulotta S, Molinaro R, Paolino D, Tassone P, Fresta M
Received 27 November 2013
Accepted for publication 29 January 2014
Published 15 May 2014 Volume 2014:9(1) Pages 2359—2372
DOI https://doi.org/10.2147/IJN.S58362
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Donato Cosco,1,5,* Cinzia Federico,1,2,* Jessica Maiuolo,1 Stefania Bulotta,1 Roberto Molinaro,1,3 Donatella Paolino,1,5 Pierfrancesco Tassone,2,4 Massimo Fresta
1Department of Health Sciences, 2Department of Experimental and Clinical Medicine, University Magna Græcia of Catanzaro, Catanzaro, Italy; 3Department of NanoMedicine, The Methodist Research Institute, Houston, TX, USA; 4Medical Oncology, Tommaso Campanella Cancer Center, Viale S Venuta, Germaneto, 5Interregional Research Center for Food Safety and Health, University of Catanzaro “Magna Græcia”, Catanzaro, Italy
*These authors contributed equally to this paper
Abstract: The aim of this study was the evaluation of the effects of two emulsifiers on the physicochemical and technological properties of low molecular weight chitosan/poly (D,L-lactide-co-glycolide) (PLGA) nanoplexes and their transfection efficiency. Nanospheres were prepared using the nanoprecipitation method of the preformed polymer. The mean diameter and surface charge of the nanospheres were investigated by photocorrelation spectroscopy. The degree of binding of the plasmid with the nanoplexes was qualitatively and quantitatively determined. MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) testing was performed using HeLa, RPMI8226, and SKMM1 cell lines. Flow cytometry and confocal laser scanning microscopy were used to determine the degree of cellular transfection and internalization of the nanoplexes into cells, respectively. The nanoplexes had a positive zeta potential, and low amounts of PLGA and poloxamer 188 showed a mean colloidal size of ~200 nm with a polydispersity index of ~0.14. The nanoplexes had suitable entrapment efficiency (80%). In vitro experiments showed that the colloidal nanodevices did not induce significant cytotoxicity. The nanoplexes investigated in this study could represent efficient and useful nonviral devices for gene delivery. Use of low amounts of PLGA and poloxamer 188 enabled development of a nanosphere able to transfect cells efficiently. These nanosystems are a helpful platform for delivery of genetic material while preserving therapeutic efficacy.
Keywords: nanoplexes, gene delivery, chitosan, poly(D,L-lactide-co-glycolide), poloxamer 188
Introduction
In recent years, gene therapy has become a promising strategy for the treatment of a number of heritable and acquired diseases. Many approaches have been used for efficient cellular gene delivery/transfection, and nonviral systems are particularly attractive because they lack the immunogenic and inflammatory side effects associated with traditional viral systems.1,2
Cationic liposomes and lipoplexes were the first positively charged delivery devices to be investigated, and were found to have a good degree of transfection but had the serious drawbacks of poor encapsulation efficiency and limited stability during storage, and were cleared rapidly from the blood.3,4 A number of cationic polymers have been investigated in order to find novel polyplexes able to assure suitable complexation and transfection, but the obtained aggregates showed a broad mean size interval and a variable shape conformation.5
Colloidal polymeric nanoparticles have been used because of their stability, their tunable physicochemical characteristics, and their potential ability to control drug leakage.6,7 Among these, poly(lactide) and poly(D,L-lactide-co-glycolide) (PLGA) nanoparticles have been widely utilized because of their excellent biodegradability, which has led to their approval by the US Food and Drug Administration for clinical use in humans.8 PLGA nanoparticles have been extensively investigated for their capacity to deliver various agents, including anticancer drugs,9,10 proteins, and peptides11 in a sustained and targeted manner. They have also demonstrated efficiency as drug delivery devices that are able to overcome obstacles, such as the blood–brain barrier12,13 and other barriers in gastrointestinal,14 nasal,15 and ocular16 tissues. Unfortunately, PLGA has a negative charge, which limits its interaction with DNA/RNA, so cationic compounds have been combined with this polymer in order to condense genetic material efficiently.17,18 For example, chitosan, a polysaccharide used widely in the pharmaceutical field because of its biocompatibility, biodegradability, safety, and mucoadhesive properties,19,20 has been used in combination with PLGA in order to increase transfection efficiency.21,22
In 2007, Nafee et al investigated the variation in physicochemical parameters of PLGA/high molecular weight chitosan nanoparticles (produced by the emulsion–diffusion–evaporation technique) as a function of the proportions of compounds contained therein. Use of PLGA 70:30 (1–2 mg/mL), poly(vinyl alcohol) (PVA, 1–2 mg/mL), and chitosan (up to 0.6 mg/mL) favored formation of systems having the smallest mean diameter yet still able to transfect efficiently.23 PLGA/PVA/chitosan nanospheres were used to entrap and deliver small interfering RNA (siRNA) material, and showed better transfection in A549 cells and a reduced cytotoxic effect compared with DOTAP/siRNA complexes.24 The same research team showed that polysorbate 80-modified PLGA nanospheres containing plasmid DNA (pDNA) enabled successful cellular transfection even in the absence of a cationic surface charge.25
PVA is the most commonly used emulsifier when formulating PLGA/poly(lactide) nanoparticles. Unfortunately, a small amount of PVA remains associated with the nanoparticles despite repeated washing, because PVA forms an interconnected network with the polymer at the interface with nanoparticles.8 The residual PVA can alter the physical properties of the nanoparticles and affect their cellular uptake, decreasing the degree of internalization.26 In this study, we produced PLGA/low molecular weight chitosan nanospheres without using PVA by exploiting the effect of Tween 80 and poloxamer 188 on their physicochemical properties. Low molecular weight chitosan was used because it enables better cell transfection than high molecular weight chitosan.27 We demonstrated that high molecular weight chitosan negatively influences the rate of interaction between siRNAs and the RNA-induced silencing complex inside the cells to a greater extent than low molecular weight chitosan/siRNA complexes.28 We also showed that PLGA nanoparticles coated with low molecular weight chitosan interact with cells in a pH-sensitive manner, enabling specific drug delivery to cells in a weakly acidic environment, with reduced opsonization and phagocytic uptake.29 Poloxamer 188 was used because of its ability to preserve and repair the cell membrane, thus increasing the structural stability of the cell.30 The ability of these nanoplexes to interact with and transfect cells was successfully investigated, along with their in vitro toxicity.
Materials and methods
Materials
The PLGA 75:25 (molecular weight 66,000–107,000), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (used for MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] tests), dimethyl sulfoxide, amphotericin B solution (250 μg/mL), phosphate saline tablets (for preparation of phosphate-buffered solution pH ~7.4), sodium dodecyl sulfate, agarose, ethidium bromide, and Hoechst (500 μg/mL) used in the experiments were purchased from Sigma Chemicals Co (St Louis, MO, USA). Tween 80 and chitosan (5 kDa, copolymer of β [1→4] linked 2-acetamido-2-deoxy-β-D-glucopyranose and 2-amino-2-deoxy-β-D-glucopyranose [degree of deacetylation 85%]) were provided by ACEF SpA (Fiorenzuola D’Arda, Piacenza, Italy). Lissamine™ Rhodamine B 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine triethylammonium salt (rhodamine DHPE) was sourced from Life Technologies (Monza, Italy). Poloxamer 188 (Pluronic® PE 6800 or Pluronic F68) was purchased from BASF (Ludwigshafen, Germany), while the cellulose membranes (Spectra/Por®, molecular weight cutoff 50,000) used in the dialysis experiments were obtained from Spectrum Laboratories Inc (Eindhoven, the Netherlands).
SKMM1 and RPMI8226 cells were provided by the Association for Cancer Research 5×1000 Network, and HeLa cells were purchased from the Istituto Zooprofilattico of Modena and Reggio Emilia. For the in vitro studies, Roswell Park Memorial Institute 1640 medium and Dulbecco’s Modified Eagle’s Medium enriched with glutamine, fetal bovine serum, and penicillin-streptomycin solution were obtained from Invitrogen (Life Technologies, Monza, Italy). The plasmid coding for the green fluorescent protein-cytomegalovirus immediate early promoter (pCMV-GFP; 3,487 base pairs) was purchased from Plasmid Factory GmbH & Co. KG (Bielefeld, Germany). All other materials and solvents used were of analytical grade (Carlo Erba, Milan, Italy).
Preparation of pCMV-GFP-loaded chitosan-PLGA nanoparticles
The nanoparticles were prepared following the nanoprecipitation method of the preformed polymer in an aqueous solution.9 The same formulation was prepared using two different emulsifier agents, ie, poloxamer 188 and Tween 80. Briefly, PLGA was dissolved in 2 mL of acetone at room temperature and added to 5 mL of acidified aqueous phase made up of MilliQ water (acetic acid 0.5% v/v) containing different amounts of chitosan, poloxamer 188, or Tween 80, which were homogenized using an Ultra-Turrax® disperser (IKA-Werke GmbH & Co. KG, Staufen im Breisgau, Germany) at 24,000 rpm for one minute and then mechanically stirred at 600 rpm for 3 hours to allow the solvent to evaporate completely (Table 1). The aqueous phase had been previously filtered using a 0.45 μm polyamide filter (Chromafil® Xtra PA-45/25; Macherey-Nagel GmbH & Co. KG, Düren, Germany) to avoid formation of chitosan aggregates. To prepare a series of formulations containing pCMV-GFP (Nano-pGFP), different amounts of plasmid were added to the aqueous solutions then stirred (Figure 1, Table 1).
Fluorescent-labeled nanoparticles were prepared by codissolving rhodamine DHPE (0.1% molar) with PLGA in the organic phase during the preparation procedure.
Physicochemical characterization and in vitro release
The mean size, size distribution, and surface charge of the different formulations were measured by photon correlation spectroscopy (Zetasizer Nano ZS; Malvern Instruments Ltd, Malvern, UK) as reported previously.31 Transmission electron microscopy was performed according to a procedure described elsewhere.32 The amount of pCMV-GFP encapsulated in the nanoparticles was spectrophotometrically determined. Briefly, nanoplexes were centrifuged and the supernatant analyzed at 268.8 nm using UV WinLab™ acquisition software (PerkinElmer GmbH, Überlingen, Germany). An empty nanoparticle formulation was used as the blank. A calibration curve was constructed to determine the amount of plasmid using the following equation:
y =49.205x−0.6173 |
where x is the concentration and y is the absorbance. The linear regression coefficient (r2) was 0.9994. Entrapment efficiency was expressed as the percentage of entrapped plasmid with respect to the amount used for preparation of the nanoplexes.
The pellet was resuspended in phosphate-buffered saline (pH 7.4) in order to investigate the release profile of pCMV-GFP from these nanosystems at 37°C using Franz-type diffusion flow cell apparatus.7 The plasmid concentration was determined using a microplate spectrophotofluorometer (Multiskan MS 6.0; Labsystems, Thermo Fisher Scientific, Waltham, MA, USA) with a Quant-It™ PicoGreen dsDNA quantitation assay reagent (Life Technologies, Grand Island, NY, USA). The integrity of the pDNA was monitored by agarose gel electrophoresis after each incubation period (see gel retardation assay). All samples were run in triplicate and the data are shown as the mean ± standard deviation.
Interaction of pDNA with nanoparticles according to gel retardation assay
The binding/condensation ability of the chitosan-PLGA nanoparticles and pCMV-GFP was determined by agarose gel electrophoresis. Briefly, the nanoplexes were centrifuged and the pellet was disrupted using sodium dodecyl sulfate (0.4% w/v) aqueous solution to determine the presence of pDNA (see Supplementary materials). Next, 10 μL of each sample was added to 2 μL of 6× loading dye, giving a total final volume of 12 μL. The complexes were then loaded onto 1% agarose gel and run with Tris-acetate-ethylenediaminetetraacetic acid buffer containing 0.5 μg/μL of ethidium bromide at 100 V for 30 minutes. The pCMV-GFP bands were visualized by irradiation with ultraviolet light using a UV-20 transilluminator (Hoefer Pharmacia Biotech Inc., San Francisco, CA, USA). Naked pCMV-GFP and blank nanoparticles were used as the controls.
Cell cultures
RPMI8226 and SKMM1 cells were maintained in suspension within tissue culture-treated flasks (75 cm2) containing Roswell Park Memorial Institute 1640 medium, and the HeLa cell line was cultured in plastic culture dishes (100 mm × 20 mm). All media were supplemented with 10% (v/v) fetal bovine serum, 100 μg/mL streptomycin, 100 IU/mL penicillin, and 250 μg/mL amphotericin B, and then incubated at 37°C in a humidified atmosphere with 5% CO2 (Forma® Series II water-jacketed CO2 incubator; Thermo Scientific, Sankt Leon-Rot, Germany). Fresh medium was substituted every 48 hours.
Evaluation of in vitro cytotoxic activity
The MTT test was performed on the above-mentioned cell lines as previously described33 to investigate the cytotoxicity of the colloidal formulations. Briefly, the cells were plated in 96-well culture dishes (2×104 cells per 0.2 mL), incubated for 24, 48, and 72 hours at 37°C in a humidified atmosphere of 5% CO2, and then treated with different concentrations of empty nanoparticles. Polymer-chitosan concentration (0.5–50 μg/mL) and time-dependent effects (24 and 48 hours of incubation) were then evaluated. The results are shown as the mean ± standard deviation of six separate experiments.
In vitro transfection
The HeLa cells were seeded in 6-well culture dishes (5×105 cells per well) for 24 hours before the experiments in order to reach 60% confluence. The medium was then replaced by an antibiotic-free medium containing the different formulations for 6 hours, ie, HpDNA and SpDNA (containing 8 μg of plasmid), empty nanoparticles (the same amount used for nanosystems containing pCMV-GFP), and calcium phosphate-pCMV-GFP (containing 8 μg of plasmid). RPMI8226 and SKMM1 myeloma cells were seeded (1×106 cells per well) in an antibiotic-free medium containing the different formulations. Untreated cells were used as the control. After 6 hours of incubation, the cells were washed and incubated for 24 and 48 hours with fresh medium.
The cells were centrifuged, the supernatant was removed, and the pellet was resuspended in prechilled phosphate-buffered saline. The cells were then transferred into a 5 mL polystyrene round-bottomed tube for analysis by flow cytometry. The mean GFP intensities of the HeLa, RPMI8226, and SKMM1 cells were recorded using a flow cytometer (FACScan™; BD Biosciences, San Jose, CA, USA). The experiments were performed in triplicate. The same fluorescence-activated cell sorting procedure was used to investigate the amount of fluorescent nanosystems taken up by the HeLa cells after 6 hours of incubation.
Confocal laser scanning microscopy
The transfection efficiency of the SpDNA formulation was investigated using HeLa cells and confocal laser scanning microscopy, as previously described.34 The cells were placed in 6-well culture plates (4×105 cells/mL) with culture medium. A sterile glass slide had been previously positioned in each well. The plates were incubated for 24 hours, and the cells were then treated with nanosystems containing pCMV-GFP (containing 8 μg of plasmid) for 6 hours. The slides were fixed after 24 hours. Each well was treated with 1 mL of Hoechst solution (1/1,000), incubated for 10 minutes, and then washed three times with 2 mL of phosphate-buffered saline. The analysis was carried out using a TCS SP2 MP laser scanning confocal microscope (Leica Microsystems, Wetzlar, Germany) at a λexc of 496 nm and a λem of 519 nm for GFP and a λexc of 405 nm and a λem of 460 nm for the Hoechst probe. A scan resolution up to 1,024×1,024 pixels and a 63× immersion oil lens was used (TCS SP2 MP laser scanning confocal microscop; Leica Microsystems, Wetzlar, Germany).
A similar experimental procedure was used to investigate the cellular uptake rate for the rhodaminated H and S formulations (Table 1) after 6 hours of incubation. Sample preparation and the analysis procedure were performed as described in the Preparation of pCMV-GFP-loaded chitosan-PLGA nanoparticles section.
Statistical analysis
The data were analyzed by one-way analysis of variance. A posteriori Bonferroni t-test was done to check the analysis of variance test. A P-value <0.05 was considered to be statistically significant. Values are reported as the mean ± standard deviation.
Results and discussion
Characterization of nanoplexes
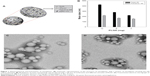
The first step was characterization of the nanoparticles in terms of their mean size, polydispersity index, and zeta potential as a function of the proportions of the different components. These parameters are fundamental for systemic administration to be possible. Table 1 shows the physicochemical characteristics of the colloids prepared with varying amounts of components. The nanospheres had a mean size of 200–500 nm and a polydispersity index of 0.15–0.4. A decrease in the amount of PLGA favored a reduction in mean particle diameter, which is consistent with the observations of Nafee et al.23 On the other hand, the formulations containing poloxamer 188 showed the smallest diameter and narrowest size distribution (Table 1, Figure 1B), probably because the steric stabilization of the particle structure was obtained as a consequence of interaction between the hydrophobic poly(ethylene oxide) center block and PLGA.35
Our findings are also in agreement with those of Zeng et al who demonstrated that a water/organic phase ratio of 2:1 v/v resulted in nanospheres with a diameter of less than 200 nm.36 It was also evident that high concentrations of chitosan favored an increase in particle size, probably as a consequence of the ability of chitosan to swell on contact with the water, which destabilizes the matrix at the time when it is becoming arranged into its final structure. The surface charges of the formulations were positive with respect to the nanospheres that did not contain chitosan as a consequence of protonation of the amino groups of the polysaccharide, thereby confirming its integration in the colloidal structure.9,37 The ideal chitosan concentration in each formulation was considered to be 0.3 mg because this favored a mean particle diameter of 150–250 nm when 6 mg of PLGA was used, and was associated with a positive zeta potential, which is useful for efficient interaction with gene material (Table 1). Further, transmission electron microscopy clearly showed that the nanospheres containing Tween 80 or poloxamer 188 were spherical in shape (Figure 1C and 1D).
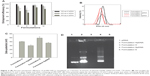
The next step was evaluation of the entrapment efficiency of the pDNA in the various nanospheres. Formulations H, J, S, and U (Table 1) were chosen for this investigation because their physicochemical parameters render them suitable for systemic gene delivery. Different amounts of pDNA were initially added for preparation of the various nanoplexes to investigate the effect of the amount of plasmid on the extent of nanosphere encapsulation. As shown in Figure 2A, the greater the initial amount of pDNA, the lower the nanosphere entrapment efficiency. Moreover, the nanoplexes showed suitable encapsulation efficiency values for their potential in vivo application. Figure 2A also shows that the nanoplexes containing poloxamer 188 allowed greater encapsulation of genetic material than the nanospheres containing Tween 80, ie, with the maximum amount of pDNA initially added (300 μg), formulations S and U showed an entrapment efficiency of over 75%. Interestingly, addition of pDNA led to a decrease in both mean size and size distribution of the various nanospheres (Table 1), ie, the SpDNA formulation had a mean size of ~150 nm and a polydispersity index of ~0.15 in comparison with formulation S, which had a mean size of 170 nm and a polydispersity index of 0.19 (Figure 2B). It is conceivable that the plasmid interacts strongly with chitosan moieties, leading to condensation of the two macromolecules and hence to greater compactness of the nanosphere matrix, with consequent shrinking of nanoparticle size.
Effective entrapment of pDNA was also demonstrated by the decreased zeta potential of the nanoplexes with respect to the empty nanospheres, ie, the surface charge decreased by ~10 mV (Figure 2C). The interaction test using gel agarose provided further evidence of the high payload of the above-mentioned nanoplexes (Figure 2D) and that the pDNA in the nanoplexes preserved the supercoiled conformation, confirming that gene material was not destabilized during the preparation process.38
Therefore, considering the aforementioned physicochemical characterization data, formulations H and S and their related nanoplexes (HpDNA and SpDNA, respectively) were used to investigate the amount of plasmid released as a function of incubation time, transfection ability, cell interaction rate, and in vitro cytotoxicity.
The pDNA release profile from the nanoplexes is another useful parameter by which to predict particle performance in the in vitro experiments. Figure 3 shows similar slow plasmid leakage from formulations H and S (~40% after 24 hours of incubation), probably as a consequence of the strong interaction between the positively charged chitosan and DNA. This trend is suitable for obtaining a good degree of cellular transfection because the pDNA remains strongly associated to the polymeric matrices that were designed to promote its uptake.
Cytotoxicity of nanospheres
The in vitro toxicity of the polymeric nanospheres in the different cell lines was investigated by MTT assay, ie, HeLa cells were used as a model of an adhesion cell line while the SKMM1 and the RPMI8226 cells were investigated as potential therapeutic targets for transfection by the nanoplexes. Novel types of gene material (miRNAs in particular) have been developed to treat myeloma and nanoplexes could be used to deliver these compounds, thereby improving their pharmacological efficacy. Evaluation of the maximum tolerated concentration of nanospheres is a fundamental prerequisite when performing transfection experiments. For this reason, different amounts of polymeric nanospheres were tested as a function of both polymer concentration and incubation time. As shown in Figure 4, formulation S did not induce significant toxicity, with the exception of the highest PLGA concentration tested, ie, only the 50 μg/mL concentration induced a reduction of cell viability of about 15%–20% after 48 hours, thus showing a cytotoxic effect at doses never used in the transfection experiments. Similar results were obtained using formulation H, demonstrating that the particles are not toxic at low polymer concentrations.
Transfection features
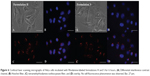
The ability of the HpDNA and SpDNA formulations to enter cells, thereby promoting expression of GFP, was investigated by fluorescence-activated cell sorting and confocal laser scanning microscopy. A calcium phosphate/pDNA formulation was used as a positive control in each experiment. As can be seen in Figure 4, an incubation time of 6 hours was sufficient to allow uptake of the nanoplexes. After 24 hours of incubation, it was possible to observe GFP protein expression in all three cell lines. As indicated by the shift in fluorescence towards the right, HeLa cells expressed the lowest degree of transfection, while the two myeloma cell lines showed a larger amount of GFP protein (Figure 5 and Supplementary materials). The SpDNA formulation gave the best results in terms of transfection efficiency, allowing a high level of GFP expression in all treated cell lines and almost full transfection of RPMI8226 cells, while the HpDNA formulation showed only limited GFP expression despite a significant shift of the fluorescence peak (Figure 5). This finding was confirmed when the cells were incubated for 48 hours following treatment with the nanoplexes (Figure 5), in that all the cell lines showed extensive fluorescent intensity as a consequence of the higher levels of protein expression. This trend might be explained by different amounts of formulations H and S being taken up by the cells. Yan et al35 demonstrated that PLGA nanoparticles containing poloxamer 188 are taken up by cells in higher concentrations with respect to the nanosystems without surfactant. Specifically, poloxamer 188 shows a higher critical insertion pressure in the lipid bilayer with respect to other poloxamers, meaning that it is a good material to be inserted in intact membranes.39
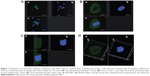
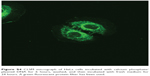
For this reason, rhodamine-labeƚed nanosystems were incubated with HeLa cells in order to investigate the degree of uptake after 6 hours of incubation by fluorescence-activated cell sorting and confocal laser scanning microscopy. Figure 6 shows good cellular localization of both nanosystems (indicated by red spots), although an appreciably greater amount of nanospheres could be obtained using poloxamer 188 as a stabilizer (formulation S), thus providing the rationale for the better fluorescence-activated cell sorting transfection profile for this formulation compared with nanosystems containing Tween 80. This trend was confirmed by fluorescence-activated cell sorting, which enabled better discrimination of the amount of fluorescent nanosystems internalized in HeLa cells after 6 hours of incubation (see Supplementary materials). Moreover, the SpDNA formulation showed characteristic green fluorescence due to expression of GFP (Figure 7). The cytosol of the transfected HeLa cells was abundantly stained 24 hours after treatment with the nanoplexes. Z-stack analysis and 180° plane rotation showed widespread green coloration, confirming a high degree of GFP protein expression in the cytosol compartment (see Supplementary materials).
Conclusion
Chitosan/PLGA nanoparticles represent a valid technological platform for gene delivery. Use of suitable amounts of poloxamer 188 instead of classical stabilizers/surfactants favored the appearance of ideal physicochemical parameters for the colloids, with high plasmid entrapment efficiency and low cytotoxicity. Further in vivo investigations of blood residence time, tissue localization, and toxicity are in progress. These nanoplexes could be a promising nanotechnology for delivering specific anti-multiple myeloma miRNAs, and are under investigation by our research team.40–42 Moreover, certain technological approaches could endow these nanoplexes with selective targeting properties against specific tumors, further improving their biodistribution and increasing the pharmacological action of the encapsulated drug(s).43 Finally, synergic action with conventional antitumor compounds could be the source of a new multidrug nanomedicine able to decrease the IC50 of single drugs and therefore their side effects.44,45
Acknowledgments
This work was financially supported by the Italian Association for Cancer Research (PI: PT. “Special Program Molecular Clinical Oncology – 5 per mille” number 9980, 2010/15) and by the PON a3_00359 (IRC-FSH) Centro Interregionale per la Sicurezza degli Alimenti e la Salute. The authors are grateful to Lynn Whitted, who revised this manuscript for English language content.
Disclosure
The authors report no conflicts of interest in this work.
References
Brannon-Peppas L, Ghosn B, Roy K, Cornetta K. Encapsulation of nucleic acids and opportunities for cancer treatment. Pharm Res. 2007;24(4):618–627. | ||
Nayerossadat N, Maedeh T, Ali PA. Viral and nonviral delivery systems for gene delivery. Adv Biomed Res. 2012;1:27. | ||
Midoux P, Breuzard G, Gomez JP, Pichon C. Polymer-based gene delivery: a current review on the uptake and intracellular trafficking of polyplexes. Curr Gene Ther. 2008;8(5):335–352. | ||
Tarahovsky YS. Cell transfection by DNA-lipid complexes – lipoplexes. Biochemistry. 2009;74(12):1293–1304. | ||
Tros de Ilarduya C, Sun Y, Düzgüneş N. Gene delivery by lipoplexes and polyplexes. Eur J Pharm Sci. 2010;40(3):159–170. | ||
Panyam J, Labhasetwar V. Biodegradable nanoparticles for drug and gene delivery to cell and tissue. Adv Drug Deliv Rev. 2003;55(3):329–347. | ||
Paolino D, Cosco D, Celano M, et al. Gemcitabine-loaded biocompatible nanocapsules for the effective treatment of human cancer. Nanomedicine (Lond). 2013;8(2):193–201. | ||
Sahoo SK, Panyam J, Prabha S, Labhasetwar V. Residual polyvinyl alcohol associated with poly(D,L-lactide-co-glycolide) nanoparticles affects their physical properties and cellular uptake. J Control Release. 2002;82(1):105–114. | ||
Cosco D, Molinaro R, Morittu VM, Cilurzo F, Costa N, Fresta M. Anticancer activity of 9-cis-retinoic acid encapsulated in PEG-coated PLGA-nanoparticles. J Drug Deliv Sci Technol. 2011;21(5):395–400. | ||
Mahapatro A, Singh DK. Biodegradable nanoparticles are excellent vehicle for site directed in-vivo delivery of drugs and vaccines. J Nanobiotechnology. 2011;9:55. | ||
Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release. 2012;161(2):505–522. | ||
Celia C, Cosco D, Paolino D, Fresta M. Nanoparticulate devices for brain drug delivery. Med Res Rev. 2010;31(5):716–756. | ||
Paolino D, Cosco D, Molinaro R, Celia C, Fresta M. Supramolecular devices to improve the treatment of brain diseases. Drug Discov Today. 2011;16(7–8):311–324. | ||
Zakeri-Milani P, Loveymi BD, Jelvehgari M, Valizadeh H. The characteristics and improved intestinal permeability of vancomycin PLGA-nanoparticles as colloidal drug delivery system. Colloids Surf B Biointerfaces. 2013;103:174–181. | ||
Keijzer C, Slütter B, van der Zee R, Jiskoot W, van Eden W, Broere F. PLGA, PLGA-TMC and TMC-TPP nanoparticles differentially modulate the outcome of nasal vaccination by inducing tolerance or enhancing humoral immunity. PLoS One. 2011;6(11):e26684. | ||
Giannavola C, Bucolo C, Maltese A, et al. Influence of preparation conditions on acyclovir-loaded poly-D,L-lactic acid nanospheres and effect of PEG-coating on ocular drug bioavailability. Pharm Res. 2003;20(4):584–590. | ||
Patil Y, Panyam J. Polymeric nanoparticles for siRNA delivery and gene silencing. Int J Pharm. 2009;367(1–2):195–203. | ||
Yuan X, Shah BA, Kotadia NK, Li J, Gu H, Wu Z. The development and mechanism studies of cationic chitosan-modified biodegradable PLGA nanoparticles for efficient siRNA drug delivery. Pharm Res. 2010;27(7):1285–1295. | ||
Ventura CA, Cannavà C, Stancanelli R, et al. Gemcitabine-loaded chitosan microspheres. Characterization and biological in vitro evaluation. Biomed Microdevices. 2011;13(5):799–807. | ||
Saranya N, Moorthi A, Saravanan S, Pandima Devi M, Selvamurugan N. Chitosan and its derivatives for gene delivery. Int J Biol Macromol. 2011;48(2):234–238. | ||
Guan XP, Quan DP, Liao KR, Tao Wang, Peng Xiang, Mai KC. Preparation and characterization of cationic chitosan-modified poly(D,L-lactide-co-glycolide) copolymer nanospheres as DNA carriers. J Biomater Appl. 2008;22(4):353–371. | ||
Niu X, Zou W, Liu C, Zhang N, Fu C. Modified nanoprecipitation method to fabricate DNA-loaded PLGA nanoparticles. Drug Dev Ind Pharm. 2009;35(11):1375–1383. | ||
Nafee N, Taetz S, Schneider M, Schaefer UF, Lehr CM. Chitosan-coated PLGA nanoparticles for DNA/RNA delivery: effect of the formulation parameters on complexation and transfection of antisense oligonucleotides. Nanomedicine. 2007;3(3):173–183. | ||
Tahara K, Yamamoto H, Hirashima N, Kawashima Y. Chitosan-modified poly(D,L-lactide-co-glycolide) nanospheres for improving siRNA delivery and gene-silencing effects. Eur J Pharm Biopharm. 2010;74(3):421–426. | ||
Tahara K, Yamamoto H, Kawashima Y. Cellular uptake mechanisms and intracellular distributions of polysorbate 80-modified poly(D,L-lactide-co-glycolide) nanospheres for gene delivery. Eur J Pharm Biopharm. 2010;75(2):218–224. | ||
Vasir JK, Labhasetwar V. Biodegradable nanoparticles for cytosolic delivery of therapeutics. Adv Drug Deliv Rev. 2007;59(8):718–728. | ||
Sato T, Ishii T, Okahata Y. In vitro gene delivery mediated by chitosan. Effect of pH, serum, and molecular mass of chitosan on the transfection efficiency. Biomaterials. 2001;22(15):2075–2080. | ||
Mao S, Sun W, Kissel T. Chitosan-based formulations for delivery of DNA and siRNA. Adv Drug Deliv Rev. 2010;62(1):12–27. | ||
Amoozgar Z, Park J, Lin Q, Yeo Y. Low molecular-weight chitosan as a pH-sensitive stealth coating for tumor-specific drug delivery. Mol Pharm. 2012;9(5):1262–1270. | ||
Moloughney JG, Weisleder N. Poloxamer 188 (p188) as a membrane resealing reagent in biomedical applications. Recent Pat Biotechnol. 2012;6(3):200–211. | ||
Paolino D, Celia C, Trapasso E, Cilurzo F, Fresta M. Paclitaxel-loaded ethosomes®: potential treatment of squamous cell carcinoma, a malignant transformation of actinic keratoses. Eur J Pharm Biopharm. 2012;81(1):102–112. | ||
Cosco D, Rocco F, Ceruti M, Vono M, Fresta M, Paolino D. Self-assembled squalenoyl-cytarabine nanostructures as a potent nanomedicine for treatment of leukemic diseases. Int J Nanomedicine. 2012;7:2535–2546. | ||
Cosco D, Bulotta A, Ventura M, et al. In vivo activity of gemcitabine-loaded PEGylated small unilamellar liposomes against pancreatic cancer. Cancer Chemother Pharmacol. 2009;64(5):1009–1020. | ||
Paolino D, Cosco D, Cilurzo F, et al. Improved in vitro and in vivo collagen biosynthesis by asiaticoside-loaded ultradeformable vesicles. J Control Release. 2012;162(1):143–151. | ||
Yan F, Zhang C, Zheng Y, et al. The effect of poloxamer 188 on nanoparticle morphology, size, cancer cell uptake, and cytotoxicity. Nanomedicine. 2010;6(1):170–178. | ||
Zeng P, Xu Y, Zeng C, Ren H, Peng M. Chitosan-modified poly(D,L-lactide-co-glycolide) nanospheres for plasmid DNA delivery and HBV gene-silencing. Int J Pharm. 2011;415(1–2):259–266. | ||
Iannone M, Cosco D, Cilurzo F, et al. A novel animal model to evaluate the ability of a drug delivery system to promote the passage through the BBB. Neurosci Lett. 2010;469(1):93–96. | ||
Liang GF, Zhu YL, Sun B, et al. PLGA-based gene delivering nanoparticle enhance suppression effect of miRNA in HePG2 cells. Nanoscale Res Lett. 2011;6:447. | ||
Haedicke A, Blume A. Interactions of Pluronic block copolymers with lipid monolayers studied by epi-fluorescence microscopy and by adsorption experiments. J Colloid Interface Sci. 2013;407:327–338. | ||
Amodio N, Di Martino MT, Foresta U, et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012;3:e436. | ||
Di Martino MT, Leone E, Amodio N, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18(22):6260–6270. | ||
Rossi M, Pitari MR, Amodio N, et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol. 2013;228(7):1506–1515. | ||
Licciardi M, Paolino D, Celia C, Giammona G, Cavallaro G, Fresta M. Folate-targeted supramolecular vesicular aggregates based on polyaspartyl-hydrazide copolymers for the selective delivery of antitumoral drugs. Biomaterials. 2010;31(28):7340–7354. | ||
Wang H, Zhao P, Su W, et al. PLGA/polymeric liposome for targeted drug and gene co-delivery. Biomaterials. 2010;31(33):8741–8748. | ||
Cosco D, Paolino D, Cilurzo F, Casale F, Fresta M. Gemcitabine and tamoxifen-loaded liposomes as multidrug carriers for the treatment of breast cancer diseases. Int J Pharm. 2012;422(1–2):229–237. |
Supplementary materials
  | Figure S1 Mean sizes (A) and correlogram (B) of formulation S after centrifugation and resuspension in sodium dodecyl sulfate aqueous solution (0.4% w/v). |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.