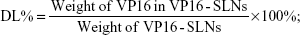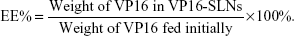Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Intracellular uptake of etoposide-loaded solid lipid nanoparticles induces an enhancing inhibitory effect on gastric cancer through mitochondria-mediated apoptosis pathway
Authors Wang J, Zhu R, Sun X, Zhu Y, Liu H, Wang S , Wang T
Received 14 March 2014
Accepted for publication 27 April 2014
Published 20 August 2014 Volume 2014:9(1) Pages 3987—3998
DOI https://doi.org/10.2147/IJN.S64103
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Jiao Wang,1 Rongrong Zhu,1 Xiaoyu Sun,1 Yanjing Zhu,1 Hui Liu,2 Shi-Long Wang1
1Research Center for Translational Medicine at East Hospital, School of Life Science and Technology, Tongji University, Shanghai, People’s Republic of China; 2Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, Shanghai, People’s Republic of China
Abstract: The objective of this study was to prepare and characterize etoposide (VP16)-loaded solid lipid nanoparticles (SLNs) and evaluate their antitumor activity in vitro. VP16-SLNs were prepared using emulsification and low-temperature solidification methods. The physicochemical properties of the VP16-SLNs were investigated by particle-size analysis, zeta potential measurement, drug loading, drug entrapment efficiency, stability, and in vitro drug-release behavior. In contrast to free VP16, the VP16-SLNs were well dispersed in aqueous medium, showing a narrow size distribution at 30–50 nm, a zeta potential value of −28.4 mV, high drug loading (36.91%), and an ideal drug entrapment efficiency (75.42%). The drug release of VP16-SLNs could last up to 60 hours and exhibited a sustained profile, which made it a promising vehicle for drug delivery. Furthermore, VP16-SLNs could significantly enhance in vitro cytotoxicity against SGC7901 cells compared to the free drug. Furthermore, VP16-SLNs could induce higher apoptotic rates, more significant cell cycle arrest effects, and greater cellular uptake in SGC7901 cells than free VP16. Moreover, results demonstrated that the mechanisms of VP16-SLNs were similar to those claimed for free VP16, including induction of cellular apoptosis by activation of p53, release of cytochrome c, loss of membrane potential, and activation of caspases. Thus, these results suggested that the SLNs might be a promising nanocarrier for VP16 to treat gastric carcinoma.
Keywords: sustained profile, gastric carcinoma, SLNs, cytochrome c, caspases
Introduction
Gastric carcinoma is one of the most common cancers and the second most frequent cause of cancer-related deaths.1 Although surgery is a preferred method of gastric carcinoma removal, it cannot remove the tissue completely and is required to be supplemented by multidrug chemotherapy and/or radiation as preferred treatment of choice. Etoposide (VP16) is an important chemotherapeutic agent that has been widely used to cure various cancers such as small-cell lung cancer, testicular cancers, and lymphomas.2–4 It inhibits the activity of topoisomerase II and causes DNA double-stranded breaks through the formation of a cleavage complex containing DNA–drug–enzyme.5 However, owing to low solubility, drug resistance, and poor bioavailability, the therapeutic efficacy of VP16 is limited.6–8 Therefore, an efficient drug delivery system is desired to overcome these drawbacks and improve the clinical therapy effects.
Nanoparticles (NPs) are emerging tools that have demonstrated great advantages in the delivery of lipophilic drugs, through: 1) dissolution of drugs in aqueous solution above their solubility limit; 2) stabilization of drugs; and 3) overcoming multidrug resistance.9,10 Besides, drug carriers in the range of 10–200 nm can passively accumulate in tumor areas through the leaky vasculature via the enhanced permeability and retention effect.11,12 Solid lipid nanoparticles (SLNs), synthesized with different types of physiological lipids, have attracted increasing attention as a promising drug delivery system because of their unique physicochemical and biological properties, including small size, good biocompatibility, the ability to protect labile compounds from chemical degradation, and the sustained release of drugs.13–15 Importantly, the physiological lipids and surfactants involved in the synthesis of SLNs have been approved by the Food and Drug Administration through the Generally Recognized as Safe rules and are used in clinical applications.16 They have been increasingly appreciated as potential drug carriers, particularly for poorly water-soluble drugs.17–20 The delivery of podophyllotoxin by SLNs for enhancing the apoptosis of HeLa cells has already been reported by our group.21
In the present study, we tried to prepare VP16 loaded SLNs (VP16-SLNs) to enhance antitumor activity. VP16-SLNs were prepared using the emulsification and low-temperature solidification method.22–25 The physicochemical properties of VP16-SLNs including particle-size analysis, zeta potential measurement, drug loading (DL), drug entrapment efficiency (EE), stability, and in vitro drug release behavior were characterized. We hypothesized that the anticancer activity of VP16-SLNs would be different from free drug; thus, the in vitro cytotoxicity as well as the effect on cell apoptosis and cell cycle with SGC7901 cells were further investigated. Finally, assessment of the mechanisms of action against SGC7901 cells was conducted. The results showed the promise of potential applications of delivering VP16 as solid lipid nanoparticles for gastric carcinoma therapy.
Materials and methods
Materials
Etoposide (≥98%), dimethyl sulfoxide, MTT formazan, and polyoxyethylene (40) stearate were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). Stearic acid, lecithin, Tween-20, and chloroform were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, People’s Republic of China). Deionized purified water was prepared in the laboratory and other reagents were all analytical grade. RPMI-1640, fetal calf serum, penicillin G, streptomycin, and trypsinase were obtained from Thermo Fisher Scientific (Waltham, MA, USA).
Preparation of VP16-SLNs
VP16-SLNs were prepared by emulsification and low-temperature solidification method. Briefly, 0.2 g stearic acid, 0.1 g lecithin, and a certain amount of VP16 were dissolved in 10 mL of chloroform in a glass flask by ultrasound to form the organic phase. The aqueous phase was composed of 0.2 g polyoxyethylene (40) stearate dissolved in 30 mL of Millipore water. The organic phase was then injected into the aqueous phase, and the mixture was stirred at 1,200 rpm at 75°C for approximately 1.5 hours until the organic solvent completely disappeared and the system volume condensed to about 5 mL. The condensed system was then quickly added to another 10 mL of cold water and stirred at 1,200 rpm for 2 hours at the temperature of 0°C–2°C. The resultant suspension was centrifuged at 20,000 rpm to remove the supernatant. The acquired precipitate was sufficiently mixed with about 30 mL of double-distilled H2O and centrifuged at 20,000 rpm. Finally, the pellet was resuspended in Millipore water, refrigerated under −80°C for 4 hours, and lyophilized in a tabletop lyophilizer. The blank carriers (SLNs) were prepared without the addition of VP16, and other experimental parameters were similar to the VP16-SLN sample.
Particle size and zeta potential analysis
Particle size and zeta potential of prepared VP16-SLNs were determined using a Malvern Nano-ZS 90 laser particle size analyzer (Malvern Instruments, Malvern, UK). All of the measurements were performed in triplicate.
Transmission electron microscopy (TEM) analysis
The morphology of VP16-SLNs was observed under a transmission electron microscope (JEOL 1230; JEOL, Tokyo, Japan). VP16-SLNs were diluted with distilled water and placed on a copper grid covered with nitrocellulose. The sample was negatively stained with 2% (w/v) sodium phosphotungstate and allowed to air-dry.
Ultraviolet-visible (UV-vis) spectral and Fourier-transform infrared (FTIR) spectral study
The presence of VP16 in the core of SLNs was examined by UV-vis spectra and FTIR. UV-vis spectra were measured on a Cary 50 UV-visible absorbance spectrophotometer (Varian, Palo Alto, CA, USA). FTIR was obtained on a Bruker Vector 22 (Bruker Corporation, Billerica, MA, USA) spectrophotometer in the range of 500–4,000 cm−1 using the standard KBr disk method (sample: KBr =1:100).
X-ray powder diffraction analysis
X-ray powder diffraction measurements were performed in order to characterize the crystallographic structure of the VP16-SLNs, VP16, and SLNs. The patterns were carried out with an X-ray diffractometer (D8 Advance; Bruker Corporation) in the range of 5°–70°. The measurements were performed at a voltage of 40 kV and 25 mA.
Determination of drug loading and entrapment efficiency
The amount of entrapped VP16 was determined by UV-vis spectrophotometer. A sample containing a standard weight of VP16-SLNs was dissolved by adding a specific amount of ethanol. After the sample was completely dissolved, the concentration of the VP16 was determined with a UV-vis spectrophotometer. The selected wavelength for VP16 measurement was 285 nm. The concentration of the VP16 was calculated according to an already-obtained calibrating curve (absorbance =0.00754[VP16]–0.00873, r =0.99991). The DL% and EE% are calculated as follows:
| (1) |
| (2) |
Sedimentation study in phosphate-buffered solution (PBS)
A total of 10 mg of VP16 was placed in a centrifugal tube of 15 mL and resuspended with 10 mL of PBS (0.01 M, pH 7.4). The tube then stood for over 6 hours after vortexing for 1 minute. Photographs of the sample were taken at 10 minutes, 3 hours, and 6 hours. Other samples were studied in the same way. Based on the DL, the same quantity of VP16 was applied to all formulations for the sedimentation study.
In vitro drug release
The in vitro release pattern of VP16 from VP16-SLNs was studied using the dialysis membrane method.26,27 In detail, 20 mg of VP16-SLNs dispersed in 12 mL PBS solution was transferred to a dialysis bag (cutoff size 14 kDa). The bag was dipped into 200 mL PBS at pH 7.4 at 37°C in a shaking water bath at 100 rpm. Samples were withdrawn from the receptor medium with replacement at indicated time intervals of up to 60 hours. The VP16 concentrations in the released samples were analyzed by UV-vis spectroscopy, as described above. The release experiments were performed in triplicate.
MTT assay
The cell viability was estimated by MTT assay.28,29 SGC7901 cells were seeded in 96-well plates at a density of 1.0×104 in 100 μL of medium per well. After 24 hours of incubation, quadruplicate wells were treated with plain SLNs, VP16, and VP16-SLNs at the concentration of 5, 10, 20 and 40 μg/mL. The plates were incubated at 37°C in 5% CO2 for 24 hours and 48 hours respectively. A control experiment was performed under the same conditions but without cell treatment. After treatment, a total of 20 μL (5 mg/mL) of MTT dye solution was added to each well before the cells were incubated for 4 hours at 37°C under a light-blocking condition. The medium was then removed and 150 μL of dimethyl sulfoxide was added into each well. Absorbance was measured at 492 nm (OD492) using an ELx800 reader (BioTek Instruments, Inc, Winooski, VT, USA), and viability of SGC7901 cells was calculated using the following equation:
| (3) |
DNA agarose gel electrophoresis
Apoptosis was assessed by the electrophoresis of extracted genomic DNA from cells.30 SGC7901 cells were seeded in 100 mm cell culture dishes at a density of 2.0×106 cells in 10 mL of medium per dish. After 24 hours of incubation, cells were treated with plain SLNs, VP16, and VP16-SLNs at a concentration of 20 μg/mL for 24 hours. A control experiment was performed under the same conditions but without cell treatment. Genomic DNA of cells was extracted using a cell genome DNA extraction kit purchased from KeyGen Biotech, Co. Ltd. (Nanjing, People’s Republic of China). Then, a certain amount of genomic DNA (4,000 ng) was mixed with loading buffer and electrophoresed in 1.8% agarose gel at 80 V for 2 hours in a mixture of tris base, acetic acid, and ethylenediaminetetraacetic acid buffer.
Hoechst 33342 staining of cells for nuclear morphology
Apoptotic nuclear morphology was assessed by Hoechst 33342 staining.31 SGC7901 cells were seeded in glass bottom dishes (In Vitro Scientific, Sunnyvale, CA, USA) at a density of 2×105 cells per dish. After 24 hours of incubation, cells were treated with a concentration (20 μg/mL) of plain SLNs, VP16, and VP16-SLNs. After 24 hours treatment, cells were washed with PBS (pH 7.4) and stained with Hoechst 33342 solution at 4°C for 30 minutes. Apoptotic nuclear morphology was observed with a confocal laser scanning microscope (FV1000 Fluoview; Olympus Corporation, Tokyo, Japan).
Apoptotic cell determination by annexin V/propidium iodide (PI) staining assay
The apoptosis analysis was also determined by annexin V-fluorescein isothiocyanate (FITC) and PI double staining.32 Briefly, SGC7901 cells were seeded in 12-well plates at a density of 1.0×106 cells per well. After 24 hours of incubation, cells were exposed to a concentration (20 μg/mL) of plain SLNs, VP16, and VP16-SLNs. After 24 hours incubation, the cells were trypsinized and collected by centrifugation. The cells were then washed twice with PBS and resuspended in 500 μL binding buffer. Then, 5 μL Annexin V-FITC and 10 μL PI solution were added after incubation in the dark at room temperature for 15 minutes, according to manufacturer’s instructions. Cell apoptosis was analyzed using a Becton–Dickinson flow cytometer (Becton, Dickinson and Company, Mountain View, CA, USA).
Cell cycle phase analysis
Cell cycle distribution based on the DNA content was determined as previously described.33 SGC7901 cells were seeded in 12-well plates at a density of 1.0×106 cells per well. After 24 hours of incubation, cells were treated with a concentration (20 μg/mL) of plain SLNs, VP16, and VP16-SLNs for another 24 hours. After being washed twice with PBS, cells were harvested and collected by centrifugation at 1,000 g for 5 minutes, followed by fixing in ice-cold 70% ethanol at 4°C overnight. Then, cells were collected by centrifugation and stained with 500 μL of PI stain solution (20 g/mL PI, 8 g/mL ribonuclease) for 30 minutes at 37°C (protected from light), followed by analysis with a Becton–Dickinson flow cytometer (Becton, Dickinson and Company). The cell distribution in phases of SubG1, G0/G1, S, and G2/M were measured and the results were calculated using Flow Jo software (version 7.6) purchased from Emerald Biotech, Co, Ltd. Hangzhou, People’s Republic of China.
Determination of mitochondrial membrane potential (Δψ)
The loss of mitochondrial membrane potential (Δψ) is a hallmark for apoptosis. It is an early event preceding phosphatidylserine externalization and coinciding with caspase activation.34,35 Mitochondria depolarization was measured using JC-1 by a switch from the red to green fluorescence. SGC7901 cells were treated with a concentration (20 μg/mL) of plain SLNs, VP16, and VP16-SLNs for 24 hours. Then, the changes in mitochondrial membrane potential (Δψ) were investigated by JC-1 apoptosis detection kit purchased from KeyGen Biotech, Co. Ltd. For confocal study, the treated cells were washed twice with PBS (pH 7.4) and incubated with JC-1 working solution. After being stained for 20 minutes at 37°C in 5% CO2 humidity, the cells were washed with 1× incubation buffer and imaged under a confocal laser scanning microscope (FV1000 Fluoview; Olympus Corporation). For fluorescence-activated cell sorting (FACS) study, the treated cells were washed twice with PBS (pH 7.4) and collected by centrifugation, followed by incubating with JC-1 working solution for 20 minutes at 37°C in 5% CO2 humidity. Then, the cells were resuspended in 500 μL 1× incubation buffer and analyzed using the Becton–Dickinson spectrophotometer (Becton, Dickinson and Company).
Cellular uptake study
For cellular uptake study, confocal microscopy was used. SGC7901 cells were seeded in glass bottom dishes (In Vitro Scientific) at a density of 2×105 cells per dish. After 24 hours of incubation, cells were treated with a concentration (20 μg/mL) of free FITC-VP16 and FITC-VP16-SLNs. After 1, 4, and 8 hours’ incubation, the cells were washed thrice with PBS (pH 7.4) to remove excess FITC-VP16-SLNs and incubated with nuclear stain 4′,6-diamidino-2-phenylindole (DAPI) for 30 minutes at room temperature. Then, the cells were viewed and imaged under a confocal laser scanning microscope (FV1000 Fluoview; Olympus Corporation) equipped with an oil immersion objective (60×; Olympus Corporation). Fluorescence images were obtained at an excitation wavelength of 488 nm (FITC) and 405 nm (DAPI) with an argon laser using FITC filter (Ex 488 nm, Em 500–600 nm).
Western blot analysis
Western blot was performed using methods provided by Mohanty and Sahoo.36 p53, cytochrome c, caspase-9, caspase-3, and caspase-7 antibodies were purchased from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). Briefly, SGC7901 cells were treated as described for the cell cycle phase analysis. Then, the cell protein lysates were extracted using a total protein extraction kit (KeyGen Biotech, Co. Ltd.). The protein content was determined with the bicinchoninic acid protein assay reagent (KeyGen Biotech, Co. Ltd.). Equal amounts of protein (20 μg) from each sample were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then electrotransferred to a polyvinylidene fluoride (PVDF) membrane (EMD Millipore, Billerica, MA, USA). Nonspecific binding was blocked by incubation with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 hour at room temperature. The blots were probed with primary antibodies against p53, cytochrome c, caspase-9, caspase-3, caspase-7, and β-actin at 1:1,000 dilutions at room temperature overnight and then washed three times with TBST. The blots were incubated with secondary antibody for 30 minutes, washed again three times with TBST, and the signal was visualized with a chemiluminescent enhanced chemiluminescent (ECL) detection system (Santa Cruz Biotechnology Inc.).
Statistical analysis
Results were expressed as mean ± standard deviation. Statistical analyses were performed by using Student’s t-test. Values of P<0.05 were considered statistically significant.
Results and discussion
As shown in Figure 1, VP16-SLNs were discrete spheres ranging from 30 nm to 50 nm in diameter as characterized by TEM (Figure 1A). Dynamic light scattering (DLS) study showed that the hydrodynamic size of VP16-SLNs was 70±1.2 nm (Figure 1C). It was reasonable that the diameter value detected via DLS was higher than that detected via TEM for the same NPs because DLS figures show the hydrodynamic radius of NPs, which are wet and extended; however, TEM represents the dry and shrunk configuration of NPs. As shown in Figure 1B, the zeta potential of the nanoparticles was -28.4 mV. The DL% and EE% were calculated as high as 36.91% and 75.42%, respectively, as listed in Table 1. The DL and EE of VP16 in the VP16-SLNs were considered optimum for further experiments.37–39 It was found that the curve of VP16-SLNs showed the characteristic absorption peak at 285 nm, which was the same as that for free VP16 (Figure 2A). As shown in Figure 2B, the spectra of free VP16, plain SLNs, and VP16-SLNs were obtained. The bands noted around 3,400 cm−1 were derived from the band vibration of O-H. The spectra of VP16 showed the following selected bands: 2,923 cm−1 (C-H stretch), 1,770 cm−1 (C=O stretch of ester bond), 1,610 cm−1 (C=O stretch of carboxyl methyl), and 1,110 cm−1 (C-O-C stretch), as well as the bands at 1,487 cm−1, which correspond to the C=C stretching in the backbone of the aromatic phenyl ring. Meanwhile, the spectra of plain SLNs showed characteristic peaks: 1,610 cm−1 (C=O stretch of carboxyl methyl), 2,930 cm−1, and 2,850 cm−1 due to C-H stretching vibration. These data suggested that VP16 was well encapsulated by SLNs, as all of the characteristic peaks for free VP16 were also identified in the VP16-SLNs. The X-ray powder diffraction study was further carried out to explore the nature of VP16 after encapsulation into SLNs (Figure 2C). The pure VP16 exhibited sharp peaks in the range of 10°–35°, which implied a highly crystalline structure; however, this characteristic was not observed in VP16-SLNs, which indicated that VP16 entrapped in the lipid core of SLNs was in amorphous or disordered-crystalline phase.40,41 This disordered-crystalline phase of VP16 inside the NPs helps in its sustained release from the VP16-SLNs. The presence of drug in crystalline form inside NPs hampers its release as such large sized molecules cannot diffuse from the small pores. However, the amorphous or disordered-crystalline phase of the drug can allow for easy diffusion through the SLNs, leading to a sustained release of the encapsulated drug.36,42,43
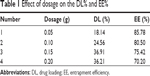  | Table 1 Effect of dosage on the DL% and EE% |
  | Figure 1 Transmission electron microscopy image (A), zeta potential distribution (B), and particle size distribution of VP16-SLNs (C). |
Figure 3 presents the photographs of free VP16, plain SLNs, and VP16-SLNs in PBS (0.01 M, pH 7.4), which were recorded after standing for 10 minutes, 3 hours, and 6 hours. It can be seen that both plain SLNs and VP16-SLNs dispersed stably in PBS, and little sedimentation of the particles was observed after standing for 6 hours. In contrast, the free VP16 added to PBS began to be precipitated and aggregated in the initial 10 minutes, and most parts of the sample still precipitated at the bottom of the tube after standing for 6 hours, even though there was a trace amount of dissolution. Therefore, the embedding of VP16 into SLNs was an effective means of improving the dispersion and stability of the drug in an aqueous suspension.
When observing the in vitro release profile, we observed a biphasic release pattern of the entrapped VP16 from the VP16-SLN formulation (Figure 4). The drug-release mechanism could belong to drug diffusion, polymer matrix swelling, and the polymer erosion or degradation. Under the experimental conditions, VP16-SLNs possessed a sustained drug release over a period of 60 hours. The initial fast drug release could be ascribed to those drugs located on or near the surface of NPs, while the slow and sustained release could be attributed to the diffusion of drug molecules through the lipid matrix of SLNs. Both burst drug release and sustained drug release are important; burst release can be useful for improving the penetration of a drug, while sustained release can be beneficial for drugs with irritation effects at high concentrations.
Figure 5A shows the viability of SGC7901 cells treated with SLNs, VP16, and VP16-SLNs at various concentrations, for 24 and 48 hours respectively. Both free VP16 and VP16-SLNs could significantly suppress SGC7901 cell proliferation in a dose- and time-dependent manner. While at the same concentration, VP16-SLNs showed significantly higher inhibition rates compared to free VP16. The average residual SGC7901 cells remained about 40% after being treated with the VP16 for 48 hours, while only 10% with VP16-SLNs. Obviously, VP16-SLNs showed higher suppression efficiency than free VP16. It is possible that good dispersity and stability of VP16-SLNs in an aqueous solution (Figure 3) could facilitate greater cellular uptake compared to free VP16. In parallel, the plain SLNs demonstrated no significant inhibitory effect.
To investigate whether VP16-SLNs induced significant DNA fragmentation in comparison to free VP16, genomic DNA was isolated from the treated SGC7901 cells and electrophoresed (Figure 5B). The results clearly showed that DNA isolated from VP16-SLN-treated cells exhibited a strong DNA smear and 180 bp DNA fragments, which were typical of apoptosis, while free VP16 showed a very faint DNA smear. Thus, it could be concluded that VP16-SLNs were more active than free VP16. The plain SLNs showed effects similar to those of the untreated control. As shown in Figure 5C, cell nuclei were round and homogeneously stained in control and plain SLN groups; however, VP16-(20 μg/mL) treated SGC7901 cells showed some marked nuclei blebbing and apoptotic bodies. The morphological changes of cell nuclei were more obvious in the VP16-SLN group, indicating that the apoptotic effects were enhanced.
Annexin-V/PI staining is a more sensitive method for detecting apoptosis. Early apoptosis was characterized by plasma membrane reorganization and was detected by positive staining for annexin V-FITC, while later stage apoptosis indicating DNA damage showed positive staining for both annexin V and PI.44 SGC7901 cells were treated with 20 μg/mL of VP16-SLNs as well as with free VP16 for 24 hours prior to FACS analysis; meanwhile, cells without any additives were set as controls. As shown in Figure 6, both early apoptotic and later apoptotic cell populations increased and VP16-SLNs showed a 13.49% higher extent of apoptosis as compared to the free VP16 drug, while the plain SLNs showed a negligible effect, which was almost the same as that of controls.
VP16 has already been shown to influence cell cycle distribution in various kinds of cells.45,46 Figure 7 shows the cell cycle histograms of SGC7901 cells treated with VP16-SLNs and free VP16 solution at identical concentrations (20 μg/mL) for 24 hours. The flow cytometry assay showed that VP16-SLNs and free VP16 solution could arrest SGC7901 cells specifically at G2/M phase cell cycle. Following exposure to free VP16 and VP16-SLNs at 20 μg/mL for 24 hours, the percentage of cells in G2/M phase were 12.35% and 17.13%, respectively, both higher than that of the control group (8.07%). These data suggested that VP16-SLNs showed more significant cell cycle arrest effect at G2/M phase in SGC7901 cells compared to free VP16 solution at the same concentration.
In fit cells, JC-1 is more concentrated in the mitochondria matrix (driven there by the Δψ), where it forms red-emitting aggregates, while, in apoptotic cells, it is in the cytosol (due to the Δψ decrease), where it exists as a green-fluorescent monomer. Accordingly, the ratio of red to green JC-1 fluorescence can be used as a sensitive measure of Δψ.47 Disruption of Δψ, a hallmark of cytochrome c translocation and the start of the apoptotic process, is indicated by a loss of red fluorescence and an increase in green fluorescence. As shown in Figure 8, both confocal microscopy and FACS study revealed that VP16, when it was entrapped by SLNs, presented a significant decrease of the mitochondrial membrane potential (Δψ). This result suggests that VP16-SLNs might induce apoptosis through the mitochondria-involved apoptosis pathway.
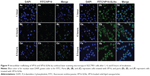
The cellular uptake of free VP16 and VP16-SLNs by SGC7901 cells is shown in Figure 9. Free VP16 (Figure 9A–C) and VP16-SLNs (Figure 9D–F) passed through the cell membrane of SGC7901 cells and assembled in the nucleus at the predetermined points of 1, 4, and 8 hours. These results demonstrate that cellular uptake in SGC7901 cells is time-dependent, and the efficient cellular uptake of VP16-SLNs is higher than that of free VP16. It could also be seen that the SLN carriers could aggregate around the nucleus (blue fluorescence) and even directly intrude into the nucleus (Figure 9Fc). These results suggest that VP16-SLNs may induce apoptosis through enhanced cellular uptake.
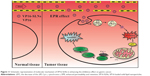
It has been demonstrated that the mitochondria-mediated apoptosis pathway played an important role in VP16-induced apoptosis.48,49 DNA damage induced by VP16 has been shown to activate p53 activity. As the activation of p53 increases, Bax synthesis and the translocation of Bax to the mitochondria induces the mitochondrial permeability transition, the event that releases cytochrome c and culminates in the death of the cells.50 It is known that cytochrome c releases from mitochondria into the cytosol, binds to the apoptotic protease-activating factor complex, and triggers the activation of cell-death signaling molecules such as caspase-9, caspase-3, and caspase-7.51 As shown in Figure 10, the results indicate that VP16-SLNs induce higher p53 activation than free VP16. Furthermore, we observed a significant increase in the marked fraction of cytochrome c released from the mitochondria of VP16-SLN-treated cells as compared to those treated with free VP16. Also, VP16-SLNs induce a greater increase in the activities of caspase-9, caspase-3, and caspase-7. Altogether, based on the results from our present study, it is clear that VP16-SLNs induce higher apoptosis of SGC7901 cells through the mitochondria-mediated apoptotic pathway (Figure 11).
Conclusion
The present study described the impact of the lipophilic drug delivery system of SLNs on improving the bioefficacy of a poorly soluble anticancer agent (VP16). The simple, economical, and reproducible preparing method could result in consistent production of smaller-sized VP16-SLNs with DL and EE. The in vitro release profile and stability data indicate that SLNs could provide sustained release of the drug and excellent physical long-term stability. VP16-SLNs also displayed an improved in vitro cytotoxicity as well as improved apoptotic effects and cellular uptake compared to free VP16 in SGC7901 cells. Also, it was demonstrated that VP16-SLNs induced a higher apoptosis ratio of SGC7901 cells through activating the mitochondria-mediated apoptosis pathway. Based on these interesting results, we can conclude that VP16-SLNs might be a good nanoformulation for the treatment of gastric carcinoma.
Acknowledgments
This work was financially supported by the 973 project of the Ministry of Science and Technology (Grant no. 2010CB912604, 2010CB933901), the International S&T Cooperation Program of China (Grant no. 0102011DFA32980), the National Natural Science Foundation of China (Grant no. 81271694, 81301157, 81301310), and the Science and Technology Commission of Shanghai Municipality (Grant no. 11411951500 and 12nm0502200).
Disclosure
The authors report no conflicts of interest in this work.
References
Shibuya K, Mathers CD, Boschi-Pinto C, Lopez AD, Murray CJ. Global and regional estimates of cancer mortality and incidence by site: II. Results for the global burden of disease. BMC Cancer. 2002;2:37. | ||
Johnson DH, Greco FA, Strupp J, Hande KR, Hainsworth JD. Prolonged administration of oral etoposide in patients with relapsed or refractory small cell lung cancer: a phase II trial. J Clin Oncol. 1990;8(10): 1613–1617. | ||
Hande KR. Etoposide: four decades of development of topoisomerase II inhibitor. Eur J Cancer. 1998;34(10):1514–1521. | ||
Sieber SM, Whang-Peng J, Botkin C, Knutsen T. Teratogenic and cytogenetic effects of some plant-derived antitumor agents (vincristine, colochicine, maytasine, VP-16-213 and VM-216) in mice. Teratology. 1978;18(1):31–47. | ||
Gantchev TG, Hunting DJ. Enhancement of etoposide (VP-16) cytotoxicity by enzymatic and photodynamically induced oxidative stress. Anticancer Drugs. 1997;8(2):164–173. | ||
Joel SP, Clark PI, Heap L, et al. Pharmacological attempts to improve the bioavailability of oral etoposide. Cancer Chemother Pharmacol. 1995;37(1–2):125–133. | ||
Lorico A, Rappa G, Finch RA, Yang D, Flavell RA, Sartorelli AC. Disruption of the murine MRP (multidrug resistance protein) gene leads to increased sensitivity to etoposide (VP-16) and increased levels of glutathione. Cancer Res. 1997;57(23):5238–5242. | ||
Qin L, Wang M, Zhu R, You S, Zhou P, Wang S. The in vitro sustained release profile and antitumor effect of etoposide-layered docuble hydroxide nanohybrids. Int J Nanomedicine. 2013;8:2053–2064. | ||
Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2(12):751–760. | ||
Wang X, Yang L, Chen ZG, Shin DM. Application of nanotechnology in cancer therapy and imaging. CA Cancer J Clin. 2008;58(2):97–110. | ||
Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent. Cancer Res. 1986;46(12):6387–6392. | ||
Maeda H, Bharate GY, Daruwalla J. Polymeric drugs for efficient tumor-targeted drug delivery based on EPR-effect. Eur J Pharm Biopharm. 2009;71(3):409–419. | ||
Müller RH, Mäder K, Gohla S. Solid lipid nanoparticles (SLN) for controlled drug delivery ± a review of the state of the art. Eur J Pharm Biopharm. 2000;50(1):161–177. | ||
Wissing SA, Kayser O, Müller RH. Solid lipid nanoparticles for parenteral drug delivery. Adv Drug Deliv Rev. 2004;56(9):1257–1272. | ||
Yang X, Liu Y, Liu C, Zhang N. Biodegradable solid lipid nanoparticle flocculates for pulmonary delivery of insulin. J Biomed Nanotechnol. 2012;8(5):834–842. | ||
Marcato PD, Durán N. New aspects of nanopharmaceutical delivery systems. J Nanosci Nanotechnol. 2008;8(5):2216–2229. | ||
Luo Y, Chen D, Ren L, Zhao X, Qin J. Solid lipid nanoparticles for enhancing vinpocetine’s oral bioavailability. J Control Release. 2006;114(1):53–59. | ||
Yuan H, Miao J, Du YZ, You J, Hu FQ, Zeng S. Cellular uptake of solid lipid nanoparticles and cytotoxicity of encapsulated paclitaxel in A549 cancer cells. Int J Pharm. 2008;348(1–2):137–145. | ||
Wang S, Chen T, Chen R, Hu Y, Chen M, Wang Y. Emodin loaded solid lipid nanoparticles: Preparation, characterization and antitumor activity studies. Int J Pharm. 2012;430(1–2):238–246. | ||
Bhushan S, Kakkar V, Pal HC, et al. Enhanced anticancer potential of encapsulated solid lipid nanoparticles of TPD: A novel triterpenediol from Boswellia serrata. Mol Pharm. 2013;10(1):225–235. | ||
Zhu RR, Qin LL, Wang M, et al. Preparation, characterization, and anti-tumor property of podophyllotoxin-loaded solid lipid nanoparticles. Nanotechnology. 2009;20(5):055702. | ||
Trotta M, Debernardi F, Caputo O. Preparation of solid lipid nanoparticles by a solvent emulsification–diffusion technique. Int J Pharm. 2003;257(1–2):153–160. | ||
Souto EB, Almeida AJ, Müller RH. Lipid nanoparticles (SLN, NLC) for cutaneous drug delivery: structure, protection and skin effects. J Biomed Nanotechnol. 2007;3(4):317–331. | ||
Li H, Zhao X, Ma Y, Zhai G, Li L, Lou H. Enhancement of gastrointestinal absorption of quercetin by solid lipid nanoparticles. J Control Release. 2009;133(3):238–244. | ||
Zhang H, Zhang FM, Yan SJ. Preparation, in vitro release, and pharmacokinetics in rabbits of lyophilized injection of sorafenib solid lipid nanoparticles. Int J Nanomedicine. 2012;7:2901–2910. | ||
Avgoustakis K, Beletsi A, Panagi Z, Klepetsanis P, Karydas AG, Ithakissios DS. PLGA-mPEG nanoparticles of cisplatin: in vitro nanoparticle degradation, in vitro drug release and in vivo drug residence in blood properties. J Control Release. 2002;79(1–3):123–135. | ||
Wang W, Zhu R, Xie Q, et al. Enhanced bioavailability and efficiency of curcumin for the treatment of asthma by its formulation in Solid lipid nanoparticles. Int J Nanomedicine. 2012;7:3667–3677. | ||
Zhong Z, Chen X, Tan W, et al. Germacrone inhibits the proliferation of breast cancer cell lines by inducing cell cycle arrest and promoting apoptosis. Eur J Pharmacol. 2011;667(1–3):50–55. | ||
Munyendo WL, Zhang Z, Abbad S, et al. Micelles of TPGS modified apigenin phospholipid complex for oral administration: preparation, in vitro and in vivo evaluation. J Biomed Nanotechnol. 2013;9(12):2034–2047. | ||
Vandita K, Shashi B, Santosh KG, Pal KI. Enhanced apoptotic effect of curcumin loaded solid lipid nanoparticles. Mol Pharm. 2012;9(12): 3411–3421. | ||
Li YB, Lin ZQ, Zhang ZJ, et al. Protective, antioxidative and antiapoptotic effects of 2-methoxy-6-acetyl-7-methyljuglone from Polygonum cuspidatum in PC12 cells. Planta Med. 2011;77(4):354–361. | ||
Qian WY, Sun DM, Zhu RR, Du XL, Liu H, Wang SL. pH-sensitive strontium carbonate nanoparticles as new anticancer vehicles for controlled etoposide release. Int J Nanomedicine. 2012;7:5781–5792. | ||
Gao JL, He TC, Li YB, Wang YT. A traditional Chinese medicine formulation consisting of Rhizoma Corydalis and Rhizoma Curcumae exerts synergistic anti-tumor activity. Oncol Rep. 2009;22(5):1077–1083. | ||
Ly JD, Grubb DR, Lawen A. The mitochondrial membrane potential (deltapsi(m)) in apoptosis; an update. Apoptosis. 2003;8(2):115–128. | ||
Han L, Du LB, Kumar A, et al. Inhibitory effects of trolox-encapsulated chitosan nanoparticles on tertbutylhydroperoxide induced RAW264.7 apoptosis. Biomaterials. 2012;33(33):8517–8528. | ||
Mohanty C, Sahoo SK. The in vitro stability and in vivo pharmacokinetics of curcumin prepared as an aqueous nanoparticulate formulation. Biomaterials. 2010;31(25):6597–6611. | ||
Ramana LN, Sethuraman S, Ranga U, Krishnan UM. Development of a liposomal nanodelivery system for nevirapine. J Biomed Sci. 2010;17:57. | ||
Fouad EA, El-badry M, Mahrous GM, Alsarra IA, Alashbban Z, Alanazi FK. In vitro investigation for embedding dextromethorphan in lipids using spray drying. Dig J Nanomater Biostruct. 2011;6(3): 1129–1139. | ||
Cui S, Yin D, Chen Y, et al. In vivo targeted deep-tissue photodynamic therapy based on near-infrared light triggered upconversion nanoconstruct. ACS Nano. 2013;7(1):676–688. | ||
Xue M, Yang MX, Zhang W, et al. Characterization, pharmacokinetics, and hypoglycemic effect of berberine loaded solid lipid nanoparticles. Int J Nanomedicine. 2013;8:4677–4687. | ||
Saxena V, Hussain MD. Polymeric mixed micelles for delivery of curcumin to multidrug resistant ovarian cancer. J Biomed Nanotechnol. 2013;9(7):1146–1154. | ||
Natu MV, de Sousa HC, Gil MH. Effects of drug solubility, state and loading on controlled release in bicomponent electrospun fibers. Int J Pharm. 2010;397(1–2):50–58. | ||
Zamani M, Morshed M, Varshosaz J, Jannesari M. Controlled release of metronidazole benzoate from poly e-caprolactone electrospun nanofibers for periodontal diseases. Eur J Pharm Biopharm. 2010;75(2):179–185. | ||
Lee MK, Lim SJ, Kim CK. Preparation, characterization and in vitro cytotoxicity of paclitaxel-loaded sterically stabilized solid lipid nanoparticles. Biomaterials. 2007;28(12):2137–2146. | ||
Chiu CC, Li CH, Ung MW, Fuh TS, Chen WL, Fang K. Etoposide (VP-16) elicits apoptosis following prolonged G2-M cell arrest in p53-mutated human non-small cell lung cancer cells. Cancer Lett. 2005;223(2):249–258. | ||
Nam C, Doi K, Nakayama H. Etoposide induces G2/M arrest and apoptosis in neural progenitor cells via DNA damage and an ATM/p53-related pathway. Histol Histopathol. 2010;25(4):485–493. | ||
Métivier D, Dallaporta B, Zamzami N, et al. Cytofluorometric detection of mitochondrial alterations in early CD95/Fas/APO-1-triggered apoptosis of Jurkat T lymphoma cells. Comparison of seven mitochondrion-specific fluorochromes. Immunol Lett. 1998;61(2–3):157–163. | ||
Park JH, Kim TH. Release of cytochrome c from isolated mitochondria by etoposide. J Biochem Mol Biol. 2005;38(5):619–623. | ||
Perkins CL, Fang G, Kim CN, Bhalla KN. The role of Apaf-1, caspase-9, and bid proteins in etoposide- or paclitaxel-induced mitochondrial events during apoptosis. Cancer Res. 2000;60(6):1645–1653. | ||
Karpinich NO, Tafani M, Rothman RJ, Russo MA, Farber JL. The Course of etoposide-induced apoptosis from damage to DNA and p53 activation to mitochondrial release of cytochromec. J Biol Chem. 2002;277(19):16547–16552. | ||
Reed JC. Cytochrome c: can’t live with it – can’t live without it. Cell. 1997;91(5):559–562. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.