Back to Journals » Breast Cancer: Targets and Therapy » Volume 6
Immunohistochemical analysis of aldehyde dehydrogenase isoforms and their association with estrogen-receptor status and disease progression in breast cancer
Authors Opdenaker L, Arnold K, Pohlig R, Padmanabhan J, Flynn D, Sims-Mourtada J
Received 16 September 2014
Accepted for publication 14 October 2014
Published 12 December 2014 Volume 2014:6 Pages 205—209
DOI https://doi.org/10.2147/BCTT.S73674
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Pranela Rameshwar
Lynn M Opdenaker,1,2 Kimberly M Arnold,1,3 Ryan T Pohlig,3,4 Jayasree S Padmanabhan,1 Daniel C Flynn,1,3 Jennifer Sims-Mourtada1–3
1Center for Translational Cancer Research, Helen F Graham Cancer Center, Christiana Care Health Services, Inc., Newark, Delaware, USA; 2Department of Biological Sciences, 3Department of Medical Laboratory Sciences, 4Biostatistics Core Facility, University of Delaware, Newark, Delaware, USA
Abstract: In many types of tumors, especially breast tumors, aldehyde dehydrogenase (ALDH) activity has been used to identify cancer stem-like cells within the tumor. The presence and quantity of these cells are believed to predict the response of tumors to chemotherapy. Therefore, identification and eradication of these cells would be necessary to cure the patient. However, there are 19 different ALDH isoforms that could contribute to the enzyme activity. ALDH1A1 and ALDH1A3 are among the isoforms mostly responsible for the increased ALDH activity observed in these stem-like cells, although the main isoforms vary in different tissues and tumor types. In the study reported here, we attempted to determine if ALDH1A1 or ALDH1A3, specifically, correlate with tumor stage, grade, and hormone-receptor status in breast-cancer patients. While there was no significant correlation between ALDH1A1 and any of the parameters tested, we were able to identify a positive correlation between ALDH1A3 and tumor stage in triple-negative cancers. In addition, ALDH1A3 was negatively correlated with estrogen-receptor status. Our data suggest that ALDH1A3 could be utilized as a marker to identify stem-like cells within triple-negative tumors.
Keywords: breast tumor, ALDH, ALDH1A1, ALDH1A3, stem-like cells, triple-negative cancer
Introduction
The aldehyde dehydrogenase (ALDH) family of enzymes are NAD(P)+-dependent enzymes that metabolize aromatic and aliphatic aldehydes. They are involved in oxidation of all-trans-retinal and 9-cis-retinal, which function in the retinoic acid (RA) cell-signaling pathway, and regulate cellular differentiation, cell-cycle progression, and apoptosis.1 High ALDH activity has been associated with self-renewal in a variety of normal and tumor tissues including those of the prostate, breast, lung, colon, cervix, and ovary.2–6 Recently, ALDH activity has become a functional marker for breast-cancer stem-like cells. When breast-cancer cells are stratified using the Aldefluor® assay, which measures the activity of ALDH using a fluorescent substrate, ALDH bright (ALDHbr) cells represent the tumor-initiating population, and are associated with drug resistance and metastasis.7,8 The presence of ALDHbr cells in the residual tumors of women who failed neoadjuvant chemotherapy predicted worse overall survival,9 indicating that these cells represent a resistant stem-like population.
There are 19 mammalian ALDH genes, belonging to eleven families and four subfamilies, whose expression and enzyme level vary among tissues.1,10–12 Of these, only a few have been shown to function in the RA signaling pathway. These include members of the ALDH1 family (ALDH1A1, ALDH1A3) as well as ALDH8A1. Immunohistochemical studies using pan-ALDH1 antibodies have shown that overexpression of ALDH1 in breast-cancer specimens is predictive of worse overall survival and is most often observed in aggressive tumors.13–15
The isoforms which contribute to the high functional activity in the Aldefluor assay are likely to be tissue specific. In normal breast tissue, the predominant isoform responsible for RA signaling is ALDH1A116 and it was predicted that, like many other tissues, ALDH1A1 activity was responsible for ALDHbr cells. However, in breast-cancer cell lines, ALDH1A3 has been shown to be the dominant isoform.17 Marcato et al published that knocking down ALDH1A1 only minimally affected the Aldefluor activity in breast cells, while the number of ALDHbr cells was significantly reduced with knockdown of ALDH1A3. They further demonstrated that protein expression of ALDH1A3, but not that of ALDH1A1, correlated with Aldefluor activity in breast-cancer cell lines and patient specimens.17
Expression of ALDH1A1 has been associated with estrogen-receptor (ER) positivity in luminal progenitors during normal breast development.18 Likewise, estrogen signaling has been shown to decrease expression of ALDH1A3 in female breast tissue.19 The relationship between the expression of ALDH1 isoforms and ER expression in breast cancer is unclear. In malignant breast tissue, overexpression of both ALDH1A1 and ALDH1A3 has been reported, however, only ALDH1A3 is reported to be positively associated with tumor grade, stage, and metastasis.17 In this report, we report on the expression of ALDH1A1 and ALDH1A3 in breast-cancer subtypes and determine their association to receptor status, grade, and progression.
Methods
Patient population
Breast-cancer tissue specimens were obtained from the biobank at the Helen F Graham Cancer Center and Research Institute under an institutional review board-approved protocol. The patient population consisted of women who underwent surgery at the institute between the years 2006 and 2013 and gave informed written consent for the samples to be biobanked. Upon surgical resection, tumors were prepared as routine formalin-fixed, paraffin-embedded blocks for sectioning. Pathological examination was performed for patient care, clinicopathologic details were recorded, and ER, progesterone receptor, and human epidermal growth factor receptor 2 (HER2) expression were determined. Hematoxylin and eosin stains of tumor sections were reviewed by a breast-cancer pathologist to determine the percentage of tumor nuclei and necrosis.
Immunohistochemical procedure
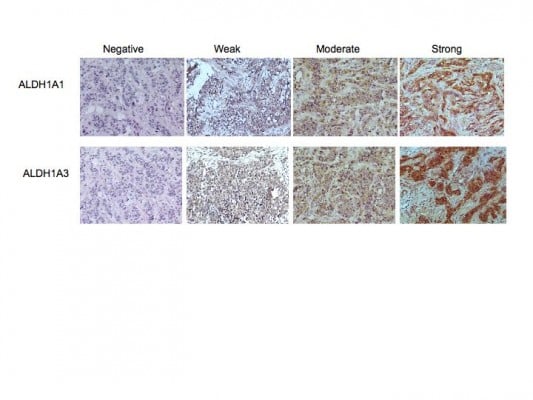
Whole 4 μm tissue sections of each specimen were analyzed by immunohistochemical staining using the LSAB+ System-HRP staining kit (Dako) according to the manufacturer’s instructions. Antibodies to ALDH1A1 (clone EP1933Y, Abcam) and ALDH1A3 (cat number ab80176, Abcam) were used at a 1:100 dilution for staining. Specificity of staining was confirmed by omission of the primary antibody and staining with an isotype-matched control antibody (Jackson Laboratories). Human liver sections served as a positive control for staining. Slides were scored as having no expression (0), or weak (1), moderate (2), or strong (3) tumor-cell staining.
Statistical analysis
Two sequential regression models were used to test the relationship between being triple negative (TN) and staining for ALDH1A1 and ALDH1A3 after adjusting for grade and stage. Assumptions for the regression analysis were met (residual normality, multicollinearity, linearity, and homoscedasticity). With the relatively small sample size it was not possible to run analysis by the full spectrum of stages (Table 1); therefore, stage was dichotomized by grouping 1A, 1B, and 2A together and 2B through 4 in a second group. The Spearman correlation matrices for the TN and ER-positive groups are presented in Table 2. Spearman was used because normality was not satisfied for every relationship using a bivariate analysis.
  | Table 1 Patient demographics and isoform immunolabeling intensities |
  | Table 2 Spearman’s correlation matrix |
Results
Patient demographics
This study comprised a cohort of 62 patients with invasive ductal carcinoma. Of these, 25 were ER positive and 37 lacked expression of ER, progesterone receptor, or HER2 (TN). A summary of the clinical data and demographics from the patient cohort is provided in Table 1. Tumor sections from 31 individuals with early stage breast cancer and from 26 with advanced stage (clinical stage ≥2B) were included in the analysis. Serial sections were immunolabeled for expression of ALDH1A1 or ALDH1A3. Slides were scored as having no expression (0), weak (1), moderate (2) or, strong (3) tumor-cell staining (Figure 1 and Table 1). Although slight stromal staining was observed in several sections for both antibodies, this staining was not included in the analysis as stromal staining of ALDH1 has been reported in normal breast stroma.20
Analysis of ALDH1A1 expression
The distribution of staining by specimen type is presented in Table 1. Of all of the breast-cancer samples, 23% (14 of 62) had no expression of ALDH1A1, 32% (20 of 62) had weak expression, 31% (19 of 62) had moderate expression, and 15% (nine of 62) had strong expression. Samples were further categorized by receptor status. Of the ER-positive tumors, 40% (ten of 25) had no expression of ALDH1A1, 24% had weak (six of 25) expression, 24% (six of 25) had moderate expression, and 12% (three of 25) had strong expression. For TN samples, 11% (four of 37) had no expression, 38% (14 of 37) had weak expression, 35% (13 of 37) had moderate expression, and 16% (six of 37) had strong expression. No association was found between the immunolabeling intensity of ALDH1A1, receptor status, grade, and stage. The base model consisting of dichotomized stage and grade was not significant (F(2,50) =1.65, P=0.20, R2=0.06, adjusted R2=0.03), nor were grade or stage individually significant. After adjusting for grade and stage, there was no significant association between receptor status and ALDH1A1 expression (F(3,49) =1.64, P=0.19, R2=0.09, adjusted R2=0.04, ΔR2=0.03). Independent of ALDH1A1 expression, receptor expression (b=0.39, SE =0.32), grade (b=0.12, SE =0.25), and stage (b=0.33, SE =0.27) were not associated (all P-values >0.05).
Analysis of ALDH1A3 expression
Analysis of ALDH1A3 staining in the all breast-cancer specimens revealed no staining in 23% (14 of 62), expression was weak in 26% (16 of 62), moderate in 31% (19 of 62), and strong in 21% (13 of 62) of samples (Table 1). Of the ER-positive samples, 44% (eleven of 25) had no expression of ALDH1A3, 36% (nine of 25) had weak expression, 12% (three of 25) had moderate expression, and 8% (two of 25) had strong expression. For TN samples, 8% (three of 37) had no expression of ALDH1A3. Expression was weak in 19% (seven of 37), moderate in 43% (16 of 37), and strong in 30% (eleven of 37) of samples. In contrast to ALDH1A1 expression, an association of ALDH1A3 to tumor grade and stage was observed for ER-positive tumors. The base model consisting of dichotomized stage and grade was significant (F(2,50) =10.90, P<0.001, R2=0.3, adjusted R2=0.28). Both grade (P=0.002) and stage (P=0.003) were also individually significant. Furthermore, in contrast to ALDH1A1, expression of ALDH1A3 was associated with hormone-receptor status. After adjusting for grade and stage, TN specimens had a significantly stronger association with ALDH1A3 than ER-positive samples (F(3,49) =12.29, P<0.001, R2=0.43, adjusted R2=0.39, ΔR2=0.13). In this model, TN receptor status (b=0.89, SE =0.27, t(49) =3.29, P=0.002) and higher stage (b=0.79, SE =0.23, t(49) =3.47, P=0.001), were both positively related to ALDH1A3. In contrast to ER-positive tumors, there was no association between ALDH1A3 and grade within the TN subtype (b=0.28, SE =0.21, t(49) =1.33, P=0.19). This is most likely due to the fact that the majority of the TN samples presented as high grade.
Discussion
In this study, we sought to examine the expression of ALDH1A family members ALDH1A1 and ALDH1A3 in breast cancer in relation to stage, grade, and hormone-receptor expression. We observed that both isoforms were significantly expressed in both ER-positive and TN tumors. However, there was no significant association between immunolabeling intensity of ALDH1A1 and tumor stage or receptor status. In contrast, ALDH1A3 was expressed at significantly higher levels in tumors that lacked expression of the ER. In both ER and TN tumors, expression of ALDH1A3 was positively correlated with tumor stage, and there was a positive association with grade in ER-positive tumors. Our studies support previous findings that indicate the expression of ALDH1A3 is inversely correlated with ER signaling,18,19 and may be predictive of metastatic potential in invasive breast cancers.17
To date, there have been many conflicting reports regarding expression of ALDH1A isoforms and their prognostic value. Some groups have found that ALDH1A is a marker of poor clinical outcome8,21 and others have found no correlation between ALDH1A expression and clinical outcome.17,22,23 Analysis of ALDH isoforms has shown that only ALDH1A3 correlates with distant metastases, disease-free survival, and overall survival in breast cancer.17 Similarly, Qiu et al found a correlation between ALDH1A3 and poor prognosis.24 However, in contrast to our findings here, they did not find a correlation between ALDH1A3 and ER status. Although our findings indicate that ALDH1A3 is associated with TN tumors, a further categorization of ER- tumors may be needed to clarify this relationship. TN breast cancers can be further classified by gene-expression profiles such as basal-like/claudin-low, mesenchymal stem-like, and luminal androgen receptor subtypes.25 The basal-like subtype is defined by the expression of cytokeratins 5 and 14 and the epidermal growth factor receptor (EGFR). Basal-like tumors often have a poor prognosis and are reported to have a higher percentage of cells that possess a stem-like phenotype.26 Recent studies have shown that expression of ALDH1A correlates with prognosis in EGFR-expressing basal-like cancers.15,26 However, specific isoforms were not examined in these studies.
Our findings indicate that ALDH1A3 correlates with stage in TN breast-cancer samples, and may be a prognostic marker for TN breast cancer. Limitations of this study include small sample size, and lack of progression-free and overall-survival information. Additionally, the TN samples in this study were not further classified into basal-like and luminal subtypes. Future work should examine the relationship of ALDH1A3 and survival within the subtypes of TN breast cancer. Additionally, the relationship between ALDH isoforms and other prognostic markers, such as Ki67, and EGFR should be determined.
Disclosure
The authors declare no conflicts of interest in this work.
References
Black W, Vasiliou V. The aldehyde dehydrogenase gene superfamily resource center. Hum Genomics. 2009;4(2):136–142. | |
Marcato P, Dean CA, Giacomantonio CA, Lee PW. Aldehyde dehydrogenase: its role as a cancer stem cell marker comes down to the specific isoform. Cell Cycle. 2011;10(9):1378–1384. | |
Liu SY, Zheng PS. High aldehyde dehydrogenase activity identifies cancer stem cells in human cervical cancer. Oncotarget. 2013;4(12): 2462–2475. | |
Ucar D, Cogle CR, Zucali JR, et al. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem Biol Interact. 2009;178(1–3): 48–55. | |
van den Hoogen C, van der Horst G, Cheung H, et al. High aldehyde dehydrogenase activity identifies tumor-initiating and metastasis-initiating cells in human prostate cancer. Cancer Res. 2010;70(12):5163–5173. | |
Huang EH, Hynes MJ, Zhang T, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009;69(8):3382–3389. | |
Reuben JM, Lee BN, Gao H, et al. Primary breast cancer patients with high risk clinicopathologic features have high percentages of bone marrow epithelial cells with ALDH activity and CD44+ CD24lo cancer stem cell phenotype. Eur J Cancer. 2011;47(10):1527–1536. | |
Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. | |
Alamgeer M, Ganju V, Kumar B, et al. Changes in aldehyde dehydrogenase-1 expression during neoadjuvant chemotherapy predict outcome in locally advanced breast cancer. Breast Cancer Res. 2014;16(2): R44. | |
Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4(6):697–720. | |
Vasiliou V, Nebert DW. Analysis and update of the human aldehyde dehydrogenase (ALDH) gene family. Hum Genomics. 2005;2(2): 138–143. | |
Sládek NE. Human aldehyde dehydrogenases: potential pathological, pharmacological, and toxicological impact. J Biochem Mol Toxicol. 2003;17(1):7–23. | |
Woodward WA, Krishnamurthy S, Lodhi A, et al. Aldehyde dehydrogenase1 immunohistochemical staining in primary breast cancer cells independently predicted overall survival but did not correlate with the presence of circulating or disseminated tumors cells. J Cancer. 2014; 5(5):360–367. | |
Currie MJ, Beardsley BE, Harris GC, et al. Immunohistochemical analysis of cancer stem cell markers in invasive breast carcinoma and associated ductal carcinoma in situ: relationships with markers of tumor hypoxia and microvascularity. Hum Pathol. 2012;44(3):402–411. | |
Morimoto K, Kim SJ, Tanei T, et al. Stem cell marker aldehyde dehydrogenase 1-positive breast cancers are characterized by negative estrogen receptor, positive human epidermal growth factor receptor type 2, and high Ki67 expression. Cancer Sci. 2009;100(6):1062–1068. | |
Petrosino JM, Disilvestro D, Ziouzenkova O. Aldehyde dehydrogenase 1A1: friend or foe to female metabolism? Nutrients. 2014;6(3): 950–973. | |
Marcato P, Dean CA, Liu RZ, et al. Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling. Mol Oncol. 2014. Epub July 24. | |
Honeth G, Lombardi S, Ginestier C, et al. Aldehyde dehydrogenase and estrogen receptor define a hierarchy of cellular differentiation in the normal human mammary epithelium. Breast Cancer Res. 2014;16(3): R52. | |
Yasmeen R, Reichert B, Deiuliis J, et al. Autocrine function of aldehyde dehydrogenase 1 as a determinant of diet- and sex-specific differences in visceral adiposity. Diabetes. 2013;62(1):124–136. | |
Ali HR, Dawson SJ, Blows FM, Provenzano E, Pharoah PD, Caldas C. Cancer stem cell markers in breast cancer: pathological, clinical and prognostic significance. Breast Cancer Res. 2011;13(6):R118. | |
Charafe-Jauffret E, Ginestier C, Iovino F, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010;16(1): 45–55. | |
Neumeister V, Agarwal S, Bordeaux J, Camp RL, Rimm DL. In situ identification of putative cancer stem cells by multiplexing ALDH1, CD44, and cytokeratin identifies breast cancer patients with poor prognosis. Am J Pathol. 2010;176(5):2131–2138. | |
Resetkova E, Reis-Filho JS, Jain RK, et al. Prognostic impact of ALDH1 in breast cancer: a story of stem cells and tumor microenvironment. Breast Cancer Res Treat. 2010;123(1):97–108. | |
Qiu Y, Pu T, Li L, et al. The expression of aldehyde dehydrogenase family in breast cancer. J Breast Cancer. 2014;17(1):54–60. | |
Ma CX, Luo J, Ellis MJ. Molecular profiling of triple negative breast cancer. Breast Dis. 2010;32(1–2):73–84. | |
Ricardo S, Vieira AF, Gerhard R, et al. Breast cancer stem cell markers CD44, CD24 and ALDH1: expression distribution within intrinsic molecular subtype. J Clin Pathol. 2011;64(11):937–946. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.