Back to Journals » International Journal of Nanomedicine » Volume 10 » Issue 1
Gold nanoparticle-based beacon to detect STAT5b mRNA expression in living cells: a case optimized by bioinformatics screen
Authors Deng D, Li Y, Xue J, Wang J, Ai G, Li X, Gu Y
Received 28 January 2015
Accepted for publication 16 March 2015
Published 28 April 2015 Volume 2015:10(1) Pages 3231—3244
DOI https://doi.org/10.2147/IJN.S81754
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Lei Yang
Dawei Deng,* Yang Li,* Jianpeng Xue, Jie Wang, Guanhua Ai, Xin Li, Yueqing Gu
Department of Biomedical Engineering, China Pharmaceutical University, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Abstract: Messenger RNA (mRNA), a single-strand ribonucleic acid with functional gene information is usually abnormally expressed in cancer cells and has become a promising biomarker for the study of tumor progress. Hairpin DNA-coated gold nanoparticle (hDAuNP) beacon containing a bare gold nanoparticle (AuNP) as fluorescence quencher and thiol-terminated fluorescently labeled stem–loop–stem oligonucleotide sequences attached by Au–S bond is currently a new nanoscale biodiagnostic platform capable of mRNA detection, in which the design of the loop region sequence is crucial for hybridizing with the target mRNA. Hence, in this study, to improve the sensitivity and selectivity of hDAuNP beacon simultaneously, the loop region of hairpin DNA was screened by bioinformatics strategy. Here, signal transducer and activator of transcription 5b (STAT5b) mRNA was selected and used as a practical example. The results from the combined characterizations using optical techniques, flow cytometry assay, and cell microscopic imaging showed that after optimization, the as-prepared hDAuNP beacon had higher selectivity and sensitivity for the detection of STAT5b mRNA in living cells, as compared with our previous beacon. Thus, the bioinformatics method may be a promising new strategy for assisting in the designing of the hDAuNP beacon, extending its application in the detection of mRNA expression and the resultant mRNA-based biological processes and disease pathogenesis.
Keywords: molecular beacon, bioinformatics, gold nanoparticle, STAT5b mRNA, visual detection
Introduction
Cancer is a distinct type of genetic disease, which is regulated by a number of signaling pathways.1 Among them, the JAK-STAT signaling pathway has been found to be highly responsible for the metastasis and proliferation of tumor cells in many human cancers, including breast cancer, lung cancer, prostate cancer, etc,2,3 and also is vital for targeted molecular cancer therapy and targeted drug screening. In the JAK-STAT signaling pathway, signal transducer and activator of transcription 5b (STAT5b) – one of the members of the STAT protein family – is an important protein, serving as a significant molecular target in the search for new biological therapeutic strategies.3 In a variety of tumor cell lines and transformed cell lines, abnormal expression and activation of STAT5b were found to be involved in the abnormal proliferation and differentiation of tumor cells. Primary patient samples of leukemic model systems have provided evidence that STAT5b plays an important role in the process of malignant transformation in acute leukemia.4,5 Activation of STAT5b increases the activity of casein promoter, which causes malignant transformation of lymphocytes.5 In addition, overexpression and constitutive activation of STAT5b have been detected in primary acute myeloid leukemia blast cells.6 Therefore, overexpression and activation of STAT5b represent a promising molecular therapeutic target in the diagnosis and study of the mechanisms of cancer.
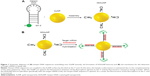
Currently, for the detection of messenger RNA (mRNA), the hairpin DNA-coated gold nanoparticle (hDAuNP) beacon has been used a facile and efficient method.7–10 Generally, the beacon consists of gold nanoparticle (AuNP; the fluorescence quencher) and hairpin DNA; the hairpin DNA is made up of a 5′ end labeled with a fluorescent dye and a stem–loop–stem oligonucleotide sequence followed by a thiol at the 3′ end.9,10 The loop oligonucleotide sequence, dominating the properties of the beacon, is designed to hybridize with a specific gene sequence, while the complementary bases modified on each side of the loop serve as the stem section to favor the hairpin structure. When the hairpin DNA is in the closed position, the fluorescence from the dye at the 5′ end is quenched due to its proximity to the AuNP (quencher) surface. When the hDAuNP beacon hybridizes specifically with the target mRNA, the hairpin DNA sequence stretches out, positioning the 5′ fluorophore away from the 3′ AuNP at a distance sufficient to give off fluorescence (Figure 1). The fluorescence recovery of the beacon is correlated with the concentration of target mRNA.
Bioinformatics use is a novel strategy to assist in the design of the complementary sequence for the construction of molecular beacon.11 In this study, we used Basic Local Alignment Search Tool (BLAST) alignment method to search for the specific STAT5b mRNA sequence with the lowest similarity to whole human and mouse genomic mRNA. BLAST is a commonly known oligonucleotide sequence comparison tool that uses a heuristic approach to construct alignments by optimizing the measurement of local similarity.12 In addition, BLAST compares protein and nucleotide sequences much faster than other methods such as the Smith–Waterman13 and Needleman–Wunsch algorithms,14 which are widely used for protein and nucleotide database search. Through this BLAST alignment method, the most specific oligonucleotide sequence that may selectively hybridize with STAT5b mRNA was chosen as the loop region of the hDAuNP beacon (its similarity ratio to other human and mouse genomic mRNA was accurately calculated). The hDAuNP beacon optimized by BLAST method possesses higher specificity (or selectivity) and sensitivity for targeted mRNA detection in living cells, as compared with our previous report based on manual selection.11
Materials and methods
Materials
Chloroauric acid (HAuCl4), sodium citrate (Na3C6H5O7), sodium borohydride (NaBH4), dithiothreitol (DTT), glutathione (GSH), sodium chloride (NaCl), magnesium chloride hexahydrate (MgCl2·6H2O), triethylamine, ethyl acetate, sodium dodecyl sulfate, Tween-20, and dimethyl sulfoxide. All chemicals were of analytical grade, purchased form Aladdin Chemistry Co Ltd (Shanghai, People’s Republic of China) and used as received.
Hairpin DNA modified with fluorescein isothiocyanate (FITC) at the 5′ end and thiol at the 3′ end was purchased from Sangon Biotech Co, Ltd (Shanghai, People’s Republic of China). Empty vector pGv113 (CON055) and positive-interference pGv113-short hairpin RNA (shRNA; 22006) were purchased from Genechem Co, Ltd (Shanghai, People’s Republic of China).
HepG-2 (human liver hepatocellular carcinoma), PC12 (rat adrenal pheochromocytoma) and MCF-7 (human breast adenocarcinoma (or Michigan Cancer Foundation-7)) cell lines were obtained from Shanghai Institutes for Biological Sciences (cell bank), Chinese Academy of Sciences. Roswell Park Memorial Institute medium 1640 (RPMI 1640) and Dulbecco’s Modified Eagle’s Medium (DMEM) were purchased from Thermo Fisher Scientific China Co, Ltd.
Optimization of STAT5b specific loop sequence with the bioinformatics method
Here, nucleotide BLAST tool was used to optimize the sequence of the specific loop region of the molecular beacon for STAT5b mRNA. Generally, 15–25 base pieces as the hybridizing loop region are suitable for the construction of the hairpin beacon. In this study, the STAT5b mRNA sequence was first obtained from The National Center for Biotechnology Information and then consecutively broken by a programmed software into 15–25 base pieces one after another with one base difference to form the sequences. Second, these bases were loaded to the nucleotide BLAST search box: (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&BLAST_PROGRAMS=megaBlast&PAGE_TYPE=BlastSearch&SHOW_DEFAULTS=on&LINK_LOC=blasthome).
Third, the sequences with similarity <70% to all genomic mRNA from human and mouse genomic databases were downloaded. Then, their secondary structures were drawn by RNAstructure software (Mathews Lab) to eliminate the sequences with inner complementary bases.
Finally, the DNA sequences complementary to the optimized specific STAT5b mRNA sequence were synthesized, serving as the loop section of hairpin DNA, while the stem bases were added. Next, RNAstructure software was used to screen the sequence with rigid hairpin loop for subsequent hDAuNP beacon. Furthermore, the beacon was introduced to assay the thermodynamic stability of the hybridization with the target mRNA.
Synthesis of STAT5b hDAuNP beacon
The AuNPs were synthesized first according to the previous method.11,15–17 Subsequently, thiol-terminated fluorescently labeled STAT5b hairpin DNA was added to the solution of AuNPs, in which the molar ratio of DNA sequence to AuNP was 110:1. After 24-hour incubation in the dark, NaCl was added to achieve the final concentration of 0.3 M and then kept at 4°C for 12 hours. At last, the reaction solution was centrifuged at 13,500× g for 20 minutes. The precipitated STAT5b hDAuNP beacon was re-suspended in pure water and stored at 4°C in the dark. Meanwhile, the supernatant was collected for determining the efficacy of conjugation of hairpin DNA onto the AuNP surface (here, the concentrations of AuNPs and the STAT5b hairpin DNA were determined by their ultraviolet [UV] absorbance at 520 nm and 260 nm, respectively).
The in vitro specificity of the as-prepared STAT5b hDAuNP beacon was determined by titration with a complementary DNA (cDNA) versus a single-base-mismatched DNA. In brief, STAT5b hDAuNP beacon was mixed with the cDNA or single-base-mismatched DNA in aqueous solution of phosphate-buffered saline (PBS) containing 0.3 mM MgCl2 and 80 mg/L bovine serum albumin at 70°C for 3 hours. After reaction, fluorescent signals at the wavelength of 520 nm were measured to assess the target selectivity of the beacon. In addition, excess DTT (0.5 M) was added to achieve complete fluorescence recovery of hDAuNP beacon.
Characterization of STAT5b hDAuNP beacon
Absorption and emission spectra of initial AuNPs and as-prepared STAT5b hDAuNP beacon were measured using a Shimadzu 3100 UV-visible-near-infrared spectrophotometer and a Shimadzu RF-5301 fluorescence spectrometer, respectively. All optical measurements were performed at room temperature. Transmission electron microscopy (TEM) images were taken on a JEOL JEM-2100 transmission electron microscope with an acceleration voltage of 200 kV. The fluorescence images of the cells were obtained with an Olympus Fluoview 300 confocal laser scanning microscope (CLSM) with 488 nm argon laser excitation. Flow cytometry (FC) analysis was performed using a BD FACSCanto flow cytometer with 488 nm excitation. In this study, the CLSM procedure was combined with FC to detect STAT5b mRNA expression in living cells, by using the optimized molecular beacon.
Qualitative imaging STAT5b mRNA in living cells
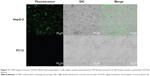
Here, HepG-2 and PC12 cell lines were used as testing and control cell lines, respectively, due to their inherent positive and negative STAT5b expression.18 These two cell lines were grown on glass-bottom wells to 50% confluence and then treated with media containing 2.0 nM STAT5b hDAuNP beacon. After 18 hours of incubation, the cells were washed three times with PBS (pH 7.4). The recovered fluorescence signals were imaged under a CLSM at excitation and emission wavelengths of 488 nm and 520 nm, respectively.
Quantitative measurement by FC
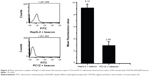
The same cell lines used in the qualitative imaging assay were used further for quantitative FC assay.19 These two cell lines were incubated with media containing 2.0 nM STAT5b hDAuNP beacon for 18 hours, rinsed thrice with PBS to remove unbound/uninternalized beacon, and then detached using trypsin. The collected cells were analyzed by FC with 488 nm excitation.
Stability and cytotoxicity of STAT5b hDAuNP beacon
Stability
For intracellular oligonucleotide beacon, endogenous nuclease degradation or endogenous biomaterial damage often occurs, which results in high background signal and/or false–positive results. Deoxyribonuclease (DNase) I and GSH are two main biodegradation- and damage-causing materials in the cytoplasm.20–23 Hence, STAT5b hDAuNP precipitate (6 nM) was resuspended in aqueous PBS solution (here, PBS aqueous solution with 0.3 mM MgCl2 and 80 mg/L bovine serum albumin was used to imitate the intracellular environment), and then DNase I (0.38 mg/L) was added. After reaction at room temperature for 1 hour, the fluorescence was measured to assess the effect of DNase I on the degradation of the beacon. Meanwhile, GSH (15 mM) was added to assess the effect of GSH on the damage to the beacon due to the possible thiol exchange. The concentrations of DNase I and GSH added refer to their common concentrations in living cells. Excess DTT (0.5 M) was used as the positive control to represent complete degradation of the hDAuNP beacon.
Cytotoxicity
The in vitro cytotoxicity of the beacon was assessed preliminarily by the colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay using HepG-2 and PC12 cells. Briefly, the cells were grown in DMEM containing 10% (v/v) fetal calf serum, 100 U/mL streptomycin, and 100 U/mL penicillin, and they were propagated in a 96-well microplate (3,000 cells per well) at 37°C for 24 hours in a humidified atmosphere containing 5% CO2. Next, the cells were incubated with various concentrations of STAT5b hDAuNP beacon. Here, the concentrations of the beacon used were set to 0 nM, 0.5 nM, 1.0 nM, 1.5 nM, 2.0 nM, and 2.5 nM. Absorption of samples at the wavelengths of 570 nm and 630 nm was measured on a microtiter plate reader. The cell viability was calculated according to the principle of MTT assay.
Assessment of STAT5b expression by reverse transcriptase–polymerase chain reaction
STAT5b expression levels in various cell lines were determined by reverse transcriptase–polymerase chain reaction (RT-PCR).11 Total RNA was extracted from freshly isolated HepG-2, PC12, or MCF-7 cell lines using RNeasy kit. RNA from each cell line was converted into cDNA with SuperScript™ III Reverse Transcriptase. Then, 1 μL cDNA was used for PCR amplification using STAT5b-specific primers:
Forward primer: 5′-CAGCGCCACGTACATG-GACCA-3′;
Reverse primer: 5′-GCGTGCGGGATCC-ACTGACTGTC-3′.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) specific primers:
Forward primer: 5′-AAGGTCGGAGTCAACGGATTT-3′;
Reverse primer: 5′-AGATGATGACCCTTTTGGCTC-3′.
β-actin-specific primers:
Forward primer: 5′-GCGTGCGGGATCCACTGACTGTC-3′;
Reverse primer: 5′-GCCGTCAGGCAGCTCGTAGC-3′.
Additionally, 5 μL of the products were used for 2% (w/v) agarose gel electrophoresis.
RNA interference of STAT5b mRNA
Bacterial (20 μL) (Escherichia coli TOP10) inoculant was seeded in Luria–Bertani medium (10 mL) containing 50 μg/mL ampicillin. Culture bottles were placed in shaking incubators (200 rpm) ensuring even air circulation. Following 10 hours of incubation at 37°C, plasmid DNA was isolated using a Plasmid Maxi kit (Qiagen, Hilden, Germany) and tested with 1% agarose gel electrophoresis. The concentration was estimated using Micro-volume Ultraviolet Spectrophotometer (Q3000, Quawell, San Jose, CA, USA).
MCF-7 cells were divided into two groups: the first group was the negative control (CON055, transfection with empty vector pGv113), and the second was the positive interference group (22006, transfection with the pGv113-shRNA). MCF-7 cells were plated in six-well plates at a density of 1×105 cells per well and incubated overnight. Cells were transfected with the empty vector and pGv113-shRNA, respectively, using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The vector backbone includes a red fluorescent protein (RFP)-coding sequence that is transcribed as part of a bicistronic transcript with the shRNA sequence, allowing the visualization of cells expressing the shRNA. RFP was observed and photographs were taken by fluorescence microscope at transfection and after 36 hours.
Statistical analysis
Significant differences were determined using the Student’s t-test, whereby differences with P<0.05 were considered significant. All data are expressed as mean ± standard error of the mean.
Results and discussion
Bioinformatics-assisted synthesis of STAT5b hDAuNP beacon
STAT5b, an important protein in the JAK-STAT signaling pathway, is responsible for the metastasis and proliferation of tumor cells.2–6,24 Therefore, visual detection of the STAT5b mRNA expression in living cells will provide insight into the mechanism of tumor progression. To achieve this scientific task, we designed a hDAuNP beacon, namely, a special hairpin DNA sequence for human STAT5b mRNA to functionalize AuNPs. Here, AuNPs serve as the fluorescence quencher and may offer efficient intracellular delivery without transfection or permeabilization reagents and an increased resistance to nuclease degradation.9–11 Meanwhile, the DNA sequence of the loop region plays a crucial role in hybridizing with the targeted mRNA, determining the sensitivity and selectivity of the beacon.8–11,25 Hence, in this study, we focused on the optimization of STAT5b specific loop sequence with the bioinformatics method.
Bioinformatics strategy has been proven to be a novel and efficient method for screening of target molecules.26–28 In this study, we used BLAST alignment method to search for specific STAT5b mRNA nucleotide sequences with the lowest similarity to all human and mouse genomic mRNAs for the construction of the molecular beacon. The detailed process has been described in the section “Optimization of STAT5b specific loop sequence” in “Materials and methods”. Next, the DNA sequence complementary to the optimized specific STAT5b mRNA sequence was synthesized, which served as the loop section of the hairpin DNA, while the complete matched base pieces – GCGAG and CTCGC – were used as the corresponding stem section. Meanwhile, a poly-A tail of ten adenosines was added to the hairpin DNA to ensure easier hybridization between STAT5b mRNA and the hDAuNP beacon in living cells. Finally, the hairpin DNA was modified with FITC at the 5′ end and thiol at the 3′ end; the 3′ thiol group enables the hairpin DNA linked to the surface of AuNP with sufficient proximity to permit an efficient quenching by the AuNPs of the fluorescence from the 5′ dye. The optimized sequences used in this study were as follows:
Hairpin DNA: 5′-FITC-(CH2)6-GCGAG-AAGCTGAA GATGGAGAGGTCG-CTCGCA(A)9SH-3′
cDNA: TTCGACTTCTACCTCTCCAGC
A single-base-mismatched DNA: TTCGACTTATACC TCTCCAGC
Figure 2A shows the UV–visible absorption spectra of AuNPs and as-prepared STAT5b hDAuNP beacon. As shown, generally, the characteristic 520 nm absorbance peak of AuNPs was retained after the covalent attachment of hairpin DNA; a little red shift and peak broadening was observed, which can be attributed to the slight aggregation for AuNP-based beacon (ultrasonication is a favorable way to eliminate this slight aggregation); the absorption peak of hairpin DNA at 260 nm was detected, indicating the formation of hDAuNP beacon. The calculated coupling ratio of hairpin DNA moieties to AuNP is approximately 90:1.
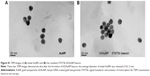
The target selectivity of the STAT5b hDAuNP beacon was assessed further by mixing it with cDNA versus single-base-mismatched DNA. After reaction, fluorescence spectra were measured and these are shown in Figure 2B. As expected, when cDNA was added, it resulted in an almost complete fluorescence recovery of STAT5b hDAuNP beacon (fluorescence intensity at 520 nm: about 71 AU) from the initial quenching state (about 2), as compared with the DTT fluorescence recovery assay (about 80 AU; excess DTT may result in a complete release of thiol-terminated FITC-labeled hairpin DNA from the AuNP surface due to a thiol–thiol exchange reaction11,16,29). However, in the presence of mismatched DNA, the solution of STAT5b hDAuNP beacon still exhibited a low fluorescent signal at 520 nm (about 5), similar to that of the initial beacon. These results demonstrate preliminarily that the STAT5b hDAuNP beacon synthesized by the bioinformatics-assisted method shows a better sensitivity (a 14-fold increase in fluorescence signal for the cDNA versus single-base-mismatched DNA), even in contrast with our recent report.11 In addition, TEM images in Figure S1 indicated that the shape and average diameter of AuNPs remained unchanged during the conjugation of hairpin DNA onto AuNP (initial AuNPs, 15±1.3 nm; the resultant STAT5b hDAuNP beacon, 15±1.5 nm).
In this section, finally, we assessed the stability and cytotoxicity of STAT5b hDAuNP beacon. The stability of the beacon against degradation by DNase I and damage by GSH was quantified by a fluorescence assay, in which excess DTT was added as a control to achieve complete fluorescence recovery (Figure S2; for an intracellular oligonucleotide beacon, DNase I and GSH are two main interfering materials in the cytoplasm.20–23). After the addition of DNase I, the fluorescent signal from STAT5b hDAuNP beacon only increased slightly; similarly, when GSH (10 mM; the intracellular concentration of GSH is within the range of 1–10 mM) was also added, the solution of the beacon exhibited low fluorescent signal. These results confirmed that the STAT5b hDAuNP beacon synthesized here is stable against degradation by DNase I and damage by GSH in the intracellular environment. Thus, the usage of this beacon for detecting STAT5b mRNA expression can avoid high background and/or false positives resulting from endogenous nuclease degradation and endogenous biomaterial damage in the intracellular environment.
The cytotoxicity of AuNPs has been proven to be size dependent.30–32 For instance, 1.4 nm AuNPs are highly cytotoxic for epithelial cells, melanoma cells, and macrophages, which may cause rapid cell death within 12 hours. In contrast, AuNPs with 15 nm in diameter are almost noncytotoxic at concentrations up to 60-fold higher than the cytotoxicity of 1.4 nm ones.31 Therefore, here, 15 nm AuNPs were synthesized and used for the construction of the hDAuNP beacon. Furthermore, we tested the cytotoxicity of STAT5b hDAuNP beacon with HepG-2 and PC12 cell lines by the MTT assay. The data in Figure S3 showed that 93%–95% of the cells exposed to the beacon were still metabolically active at concentrations up to 2.5 nM. Thus, the STAT5b hDAuNP beacon ranging from 0.5 nM to 2.5 nM did not remarkably alter the cell viability, supporting the safety and effectiveness of the beacon for in vivo imaging.
Detection of STAT5b mRNA in living cells with the optimized beacon
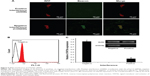
The experimental results described above have demonstrated preliminarily that the hDAuNP beacon optimized by the bioinformatics method possesses favorable sensitivity and specificity to STAT5b mRNA, besides high stability and low cytotoxicity. Next, the hDAuNP beacon was used to detect STAT5b mRNA expression in living cells. Here, STAT5b expression-positive HepG-2 and STAT5b expression-negative PC12 cell lines were used as testing and control groups, respectively.18,33,34 Qualitative assessment was performed by CLSM imaging (Figure 3). As shown, after treatment with the hDAuNP beacon, HepG-2 cells all displayed a strong fluorescent signal, while PC12 cells did not exhibit any obvious fluorescent signal. Next, we used FC to quantitatively measure the fluorescent signals in the two cell lines (Figure 4). In general, HepG-2 cells treated with STAT5b hDAuNP beacon have a stronger fluorescent signal, about 3.1-fold increase as compared with that of PC12 cells, similar to the experimental observation obtained by CLSM imaging.
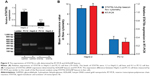
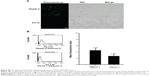
To determine the underlying mechanism for these findings, we measured STAT5b mRNA expression levels in HepG-2 and PC12 cell lines by RT-PCR, in which GAPDH mRNA was measured as the control (Figure 5). It was found that HepG-2 cells have a higher STAT5b mRNA expression level than that in PC12 cells. That is, HepG-2 and PC12 cell lines can be considered to be STAT5b expression-positive and STAT5b expression-negative, respectively. The expression difference in STAT5b mRNA between the two cell lines measured by RT-PCR was well coincident with the results from the cell imaging and FC assay. As such, the experimental results in Figures 3 and 4 should be attributed to the difference in STAT5b mRNA expression level and the resultant various degrees of fluorescence recovery for endocytic (internalized) hDAuNP beacons in these two cell lines.
Next, we verified further the specificity of the hDAuNP beacon to STAT5b mRNA by transfecting anti-STAT5b shRNAs into STAT5b expression-positive MCF-7 cells (Figure S4). As expected, after transfection, the fluorescence of RFP could be observed in the MCF-7 cells, suggesting that the vectors of anti-STAT5b shRNAs had been successfully transfected into the cells (Figure S5). Subsequently, the hDAuNP beacon was used to treat the cells before and after transfection (Figure 6). In the cells with positive interference, anti-STAT5b shRNAs did cause a significant reduction (from 0.92 to 0.24 the ratio of STAT5b mRNA expression to β-actin mRNA expression [the control]) in the STAT5b mRNA expression level after transfection and a resultant decrease in the fluorescence signal, in comparison with negative-interference cells. And the data from the FC assay were consistent with the assessment of STAT5b expression in cells by RT-PCR (the negative-interference cells treated with beacons showed 3.5-fold stronger fluorescence compared to the positive-interference cells, while the corresponding expression difference in STAT5b mRNA examined by RT-PCR is about 3.8-fold).
Finally, we wanted to preliminarily compare this STAT5b hDAuNP beacon optimized by the bioinformatics method with our previous hDAuNP beacon (or beacon 2) for the detection of targeted mRNA in living cells. The experimental result is shown in Figure S6. When the previous STAT5b hDAuNP beacon 2 was used, similar results were observed, while for the FC assay, only 1.7-fold increase in fluorescence signal was detected for STAT5b expression-positive HepG-2 and STAT5b expression-negative PC12 cell lines. Therefore, the sensitivity of STAT5b hDAuNP beacon synthesized herein was higher than that for the beacon of our previous report. These results in Figures 3–5 and S6 confirm that the bioinformatics method might be an efficient strategy for improving the specificity of the loop region sequence of the hDAuNP beacon to target mRNA.
Conclusion
The hDAuNP beacon is a versatile nanoplatform for visual detection of specific mRNAs in living cells. It has been proven that its specificity and sensitivity can be improved by a BLAST bioinformatics strategy, as outlined in this work, by the combined characterizations of optical techniques, FC assay, cell microscopic imaging, and RT-PCR technique. This achievement will extend further the potential application of the hDAuNP beacon technique in mRNA-related biological and medical processes in living cells, which even might enable the early diagnosis and the discovery of new molecular targets.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (grant numbers 81371627, 81220108012, 61335007, 81371684, 81171395, and 81328012).
Disclosure
The authors report no conflicts of interest in this work.
References
Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–141. | ||
Yoshikawa H, Matsubara K, Qian GS, et al. SOSC-1, a negative regulator of the JAK-STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet. 2001;28:29–35. | ||
Buettner R, Mora LB, Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin Cancer Res. 2002;8:945–954. | ||
Lin TS, Mahajan S, Frank DA. STAT signaling in the pathogenesis and treatment of leukemias. Oncogene. 2000;19:2496–2504. | ||
Birkenkamp KU, Geugien M, Lemmink HH, Kruijer W, Vellenga E. Regulation of constitutive STAT5 phosphorylation in acute myeloid leukemia blasts. Leukemia. 2001;15:1923–1931. | ||
Spiekermann K, Bagrintseva K, Schwab R, Schmieja K, Hiddemann W. Overexpression and constitutive activation of FLT3 induces STAT5 activation in primary acute myeloid leukemia blast cells. Clin Cancer Res. 2003;9:2140–2150. | ||
Dubertret B, Calame M, Libchaber AJ. Single-mismatched detection using gold-quenched fluorescent oligonucleotides. Nat Biotech. 2001; 19:365–370. | ||
Rosi NL, Giljohann DA, Thaxton CS, Lytton-Jean AK, Han MS, Mirkin CA. Oligonucleotide-modified gold nanoparticles for intracellular gene regulation. Science. 2006;312:1027–1030. | ||
Harry SR, Hicks DJ, Amiri KI, Wright DW. Hairpin DNA coated gold nanoparticles as intracellular mRNA probes for the detection of tyrosinase gene expression in melanoma cells. Chem Commun. 2010;46: 5557–5559. | ||
Jayagopal A, Halfpenny KC, Perez JW, Wright DW. Hairpin DNA-functionalized gold colloids for the imaging of mRNA in live cells. J Am Chem Soc. 2010;132:9789–9796. | ||
Xue JP, Shan LL, Chen HY, et al. Visual detection of STAT5B gene expression in living cell using the hairpin DNA modified gold nanoparticle beacon. Biosens Bioelectron. 2013;41:71–77. | ||
Altschul SF, Koonin EV. Iterated profile searches with PSI-BLAST: a tool for discovery in protein databases. Trends Biochem Sci. 1998; 23:444–447. | ||
Liu YC, Douglas LM, Bertil S. CUDASW++: optimizing Smith-Waterman sequence database searches for CUDA-enabled graphics processing units. BMC Res Notes. 2009;2:73. | ||
Day R. Examining the validity of the Needleman-Wunsch algorithm in identifying decision strategy with eye-movement data. Decis Support Syst. 2010;49:396–403. | ||
Kalmodia S, Harjwani J, Rajeswari R, et al. Synthesis and characterization of surface-enhanced Raman-scattered gold nanoparticles. Int J Nanomedicine. 2013;8:4327–4338. | ||
Deng DW, Zhang DY, Li Y, Achilefu S, Gu YQ. Gold nanoparticles based molecular beacons for in vitro and in vivo detection of the matriptase expression on tumor. Biosens Bioelectron. 2013;49: 216–221. | ||
Elia P, Zach R, Hazan S, Kolusheva S, Porat Z, Zeiri Y. Green synthesis of gold nanoparticles using plant extracts as reducing agents. Int J Nanomedicine. 2014;9:4007–4021. | ||
Bromberg J, Darnell JE Jr. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 2000;19: 2468–2473. | ||
Fleisher TA, Dorman SE, Anderson JA, Vail M, Brown MR, Holland SM. Detection of intracellular phosphorylated STAT-1 by flow cytometry. Clin Immunol. 1999;90:425–430. | ||
Demers LM, Mirkin CA, Mucic RC, et al. A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal Chem. 2000;72:5535–5541. | ||
Wang ZX, Ma LN. Gold nanoparticle probes. Coordin Chem Rev. 2009; 253:1607–1618. | ||
Sies H. Glutathione and its role in cellular functions. Free Radic Biol Med. 1999;27:916–921. | ||
Anderson ME. Glutathione: an overview of biosynthesis and modulation. Chem Biol Interact. 1998;112:1–14. | ||
Moser C, Ruemmele P, Gehmert S, et al. STAT5b as molecular target in pancreatic cancer – inhibition of tumor growth, angiogenesis, and metastases. Neoplasia. 2012;14:915–925. | ||
Carpi S, Fogli S, Giannetti A, et al. Theranostic properties of a survivin-directed molecular beacon in human melanoma cells. PLoS One. 2014; 9:e114588. | ||
Li J, Liu S, Koh JJ, et al. A novel fragment based strategy for membrane active antimicrobials against MRSA. Biochim Biophys Acta. 2015; 1848:1023–1031. | ||
Grover S, Dhanjal JK, Goyal S, Grover A, Sundar D. Computational identification of novel natural inhibitors of glucagon receptor for checking type II diabetes mellitus. BMC Bioinformatics. 2014; 15(suppl 16):S13. | ||
Jayadeepa RM, Ray A, Naik D, Sanyal DN, Shah D. Review and research analysis of computational target methods using BioRuby and in silico screening of herbal lead compounds against pancreatic cancer using R programming. Curr Drug Metab. 2014;15:535–543. | ||
Hoypierres J, Dulong V, Rihouey C, Alexandre S, Picton L, Thébault P. Two methods for one-point anchoring of a linear polysaccharide on a gold surface. Langmuir. 2015;31:254–261. | ||
Yang Y, Nan J, Hou J, et al. Cytotoxicity of gold nanoclusters in human liver cancer cells. Int J Nanomedicine. 2014;9:5441–5448. | ||
Pan Y, Neuss S, Leifert A, et al. Size-dependent cytotoxicity of gold nanoparticles. Small. 2007;3:1941–1949. | ||
Chung E, Nam SY, Ricles LM, Emelianov SY, Suggs LJ. Evaluation of gold nanotracers to track adipose-derived stem cells in a PEGylated fibrin gel for dermal tissue engineering applications. Int J Nanomedicine. 2013;8:325–336. | ||
Bonder MJ, Kasela S, Kals M, et al. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics. 2014;15:860. | ||
Goldshmit Y, Walters CE, Scott HJ, Greenhalgh CJ, Turnley AM. SOCS2 induces neurite outgrowth by regulation of epidermal growth factor receptor activation. J Biol Chem. 2004;279:16349–16355. |
Supplementary materials
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.