Back to Journals » Infection and Drug Resistance » Volume 7
Genotypes and phenotypes of Shiga toxin-producing Escherichia coli (STEC) in Abeokuta, Southwestern Nigeria
Authors Olowe OA , Aboderin B, Idris O, Mabayoje V, Opaleye O, Adekunle OC, Olowe RA, Akinduti PA , Ojurongbe O
Received 15 April 2014
Accepted for publication 17 June 2014
Published 15 October 2014 Volume 2014:7 Pages 253—259
DOI https://doi.org/10.2147/IDR.S66268
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Olugbenga Adekunle Olowe,1 Bukola W Aboderin,1,2 Olayinka O Idris,3 Victor O Mabayoje,4 Oluyinka O Opaleye,1 O Catherine Adekunle,1 Rita Ayanbolade Olowe,1 Paul Akinniyi Akinduti,5 Olusola Ojurongbe1
1Department of Medical Microbiology and Parasitology, College of Health Sciences, Osogbo, Osun State, Nigeria; 2Medical Microbiology Unit, Pathology Department, Federal Medical Centre, Abeokuta, Nigeria; 3Department of Microbiology, College of Sciences, Afe Babalola University, Ado Ekiti, Nigeria; 4Department of Haematology, College of Health Sciences, Ladoke Akintola University, Osogbo, Osun State, Nigeria; 5Department of Medical Microbiology and Parasitology, Olabisi Onabanjo University, Ago-Iwoye, Ogun State, Nigeria
Purpose: To characterize the prevalence of hemolytic Shiga toxin-producing Escherichia coli (STEC) with a multidrug-resistant pattern in different age groups in Abeokuta, Nigeria.
Methods: Nonrepetitive E. coli isolates were collected from 202 subjects with or without evidence of diarrhea. Each isolate was biochemically identified and antimicrobial susceptibility testing was performed using the disk diffusion method. A sorbitol fermentation test of all the E. coli isolates was done and the minimum inhibitory concentration of suspected STEC was measured by the standard broth microdilution method to determine antibiotic resistance. The genotypes of stx1, stx2, and hlyA were determined by polymerase chain reaction assay.
Results: The majority of subjects were aged ≥40 years (41.6%) and were female (61.9%). Of the 202 subjects, 86.1% had STEC isolates (P<0.05). A high rate of STEC isolates resistant to amoxicillin (90.6%), cefotaxime (77.7%), and cefuroxime (75.7%) was observed. Resistance to amoxicillin, gentamicin, and cefotaxime was demonstrated with a minimum inhibitory concentration >16 µg/mL in 13.9%, 11.4%, and 10.4% of the isolates, respectively. The prevalence of stx1, stx2, and hlyA was 13.9%, 6.9%, and 2.0%, respectively; 5.5% of stx1 were in the 0–10-year-old age group, 3.5% of stx2 were aged ≥40 and above, and 1.0% of the hlyA isolates were in the 0–10-year-old age group.
Conclusion: The prevalence of virulent STEC is a public health concern. The use of polymerase chain reaction assay should aid quick detection of this virulent serotype and help curb the severe epidemic of human diseases associated with STEC infections.
Keywords: STEC, resistance, stx1, stx2, hlyA, Nigeria
Introduction
Enteric infections continue to be one of the foremost public health problems worldwide, with over 1.5 million deaths occurring each year in developing countries.1–6 Escherichia coli is a ubiquitous bacterial organism that is found in a wide variety of places, including the human intestine, where it can lead to diarrheal disease1 and a range of extraintestinal infections.7 The increasing emergence of pathogenic E. coli, with its multiple toxigenic characteristics, has been previously reported and is considered a serious threat to public health.8 As different hosts and tissues present an alternative access to bacterial virulence factors, pathogenic E. coli often exhibits host and tissue specificity, different levels of colonization and pathogenicity, and a high degree of virulence in the host.9
The use of phenotypic characterization of E. coli for epidemiological studies of its spread and prevalence have generally relied on traditional use of the biochemical activities of the organism using selective media and specific fermentation growth characteristics,10 but these limit proper identification. In the last few decades, serological tests have been used to identify the pathogenic types based on its somatic antigen, (ie, O-antigen) and have been defined by the composition of the sugar residues in the repeating unit of outer membrane lipopolysaccharides, which can cause cross-reactivity of different genera of the bacteria isolates.11
Shiga toxin-producing E. coli (STEC) is a known zoonotic food and water-borne pathogen associated with diarrhea, hemorrhagic colitis (HC), hemolytic uremic syndrome (HUS), and renal failure, particularly in children; and it can cause attaching and effacing properties in diarrhea cases.2,11,12 The heterogeneous spread of both stx1 and stx2 has been related to the ability of STEC strains to cause serious disease in humans. Only stx2-producing STEC strains have been shown to be more commonly associated with serious human diseases compared with strains that produce stx1 alone or both stx1 and stx2. This is possibly because the level of transcription of stx2 in vivo is higher than that of stx1.13 In addition to the ability of many STEC strains to induce severe infection, they possess plasmid-encoded enterohemolysin of STEC (hlyA), which is often associated with severe clinical disease in raw meat, pigs, and humans.3,14–17
Ruminants, especially cattle,1 pigs,14 and sheep,3 unspecified meat and sheep meat,15 young cattle,16 and unpasteurized milk from cattle12 have been reported to constitute a vast range of STEC. It is not surprising that humans most frequently become infected with STEC by ingestion of contaminated food or water or by direct contact with animals, resulting in sporadic cases of disease or outbreaks involving up to several thousand individuals.4,18 Sources of infection include meat (especially undercooked beef hamburgers), ready-to-eat sausages, raw milk, cheese, unpasteurized apple cider and juice, lettuce, cantaloupes, alfalfa sprouts, radish sprouts, drinking water, water for bathing, and contact with animals.1
The prevalence of STEC has led to high mortality rates, mostly in poorly developed regions in Nigeria.19 It largely occurs via the fecal–oral route and usually results in symptoms ranging from mild uncomplicated diarrhea to severe bloody diarrhea that can progress to HC, HUS, and sometimes death.20 This is further complicated by an increased resistance to antibiotics among urinary tract E. coli strains.21 The rapid spread of resistant STEC serotypes is fast becoming a threat to public health and increased morbidity, probably due to the consumption of unhygienic food or animal food products.
The use of molecular characterization can further identify the virulent STEC strains prevalent in this community that have not usually been considered as one of the major causes of increasing colitis among children, the elderly, and immune-compromised individuals. Therefore, molecular characterization of the hemolytic STEC causing severe diarrhea was studied in Southwestern Nigeria to proffer methods of preventing the scourge.
Materials and methods
Sampling population
Nonrepetitive E. coli isolates were collected from 202 subjects with or without evidence of diarrhea attending the public health facilities in Abeokuta, Southwestern Nigeria. This a cosmopolitan town located within the rain forest belt at 6.2–7.8 degrees north latitude and 3.0–5.0 degrees east longitude, covering a land area of about 16,409.26 km2, and with population of about 3.7 million people.22 Informed consent was obtained from each individual participant, and they were assured of confidentiality in view of the intricacy and sensitivity of the study.
Isolation and biotyping
All of the E. coli samples collected were cultured on MacConkey agar with salt and eosin methylene blue agar and incubated at 37°C for 18–24 hours. Eosin methylene blue agar was used to subculture and purify the E. coli, while brain heart infusion agar (Thermo Fisher Scientific, Waltham, MA, USA) containing 7% human blood was used to test for their hemolytic pattern. Biochemical tests were performed to further characterize the suspected E. coli isolates according to the World Health Organization’s manual for laboratory investigation of acute enteric infections.5
Antimicrobial susceptibility testing
The susceptibility pattern of each E. coli isolate was tested against commonly used antibiotics by the disk diffusion method on Mueller Hinton agar according to Bauer et al.23 The following antibiotic disks were used: ampicillin (10 μg), amoxicillin/clavulanic acid (20 μg/10 μg), cefotaxime (30 μg), ceftazidime (30 μg), cefuroxime (30 μg), gentamicin (10 μg), and ciprofloxacin (5 μg). Pure isolates of 0.5 McFarland was spread on Mueller Hinton agar and the antibiotic disks were incubated at 37°C for 18–24 hours. The inhibition zones were measured and interpreted as sensitive, intermediate, or resistant according to Clinical and Laboratory Standards Institute guidelines.24
Sorbitol fermentation test
Each E. coli isolate was subcultured on sorbitol MacConkey agar. They were incubated at 37°C for 18–24 hours and the isolates showing pale coloration, ie, sorbitol nonfermenting isolates, were presumed to be STEC.
Minimum inhibitory concentration (MIC)
The standard broth microdilution method was used to determine the MIC of the obtained STEC strains. All of the presumed STEC isolates were tested against the following antibiotic dilution ranges in microtubes: ceftazidime (1–64 μg/mL), ofloxacin (0.5–64 μg/mL), cotrimoxazole (0.5–64 μg/mL), gentamicin (0.5–64 μg/mL), cefuroxime (1–64 μg/mL), cefotaxime (1–64 μg/mL), and amoxicillin (0.25–64 μg/mL), using a broth culture of 0.5 McFarland followed by incubation at 37°C in ambient air for 24 hours. Turbid wells were indicated to have growth, while clear wells were identified as having no growth after incubation. The MIC of each antibiotic to the resistant organism was noted as the highest dilution showing no growth. The respective MIC of each isolate was determined and interpreted according to Clinical and Laboratory Standards Institute guidelines.24
Genomic analysis
Chromosomal DNA of the overnight broth culture of the isolates was obtained using the cetyl trimethylammonium bromide extraction method described by Minas et al.25 The obtained DNA pellets were resuspended in 200 μL sterile water and quantified by gel electrophoresis photo-documentation.
Polymerase chain reaction (PCR) assay for stx1
The stx1 amplification was carried out in a final reaction volume of 20 μL using 10 μL 2× PCR Master Mix containing 1.5 μL MgCl2 (25 mM), 0.5 μL deoxyribonucleotide triphosphates (10 mM; deoxyadenosine triphosphate, deoxyguanosine triphosphate, deoxycytidine triphosphate, deoxythymidine triphosphate), 1 μL stx1 (2.5 μM) forward primer (5′-ACACTGGATGATCTCAGTGG-3′) and reverse primer (5′-CTGAATCCCCCTCCTTATG-3′), 1 μL stx1 (2.5 μM), 0.2 μL Taq polymerase enzyme, ≤20 μL nuclease-free water, and 2.0 μL of each extracted chromosomal DNA. The final reaction mixture was placed in a thermal cycler block (ATC 401; Nyx Technik, San Diego, CA, USA). The amplification reaction was carried out in 30 repeated temperature changes called cycles of initialization - temperature of 94°C for 2 minutes, denaturation reaction of 94°C for 30 seconds which causes melting of DNA template and primers then followed by 48°C for 30 seconds annealing of the primers to the single-stranded DNA template, elongation at optimum temperature activity of 72°C for 60 seconds to produce complementary multiple DNA.26–28
PCR amplification for stx2
The stx2 amplification was carried out in a final reaction volume of 20 μL using 10 μL 2× PCR Master Mix, containing 1.5 μL MgCl2 (25 mM), 0.5 μL deoxyribonucleotide triphosphates (10 mM; deoxyadenosine triphosphate, deoxyguanosine triphosphate, deoxycytidine triphosphate, deoxythymidine triphosphate), 1 μL stx2 (2.5 μM) forward primer (5′-CCATGACAACGGACAGCAGTT-3′) and reverse primer (5′-CCTGTCAACCGGTGAGCGCACTTTC-3′), 1 μL stx2 (2.5 μM), 0.2 μL Taq polymerase enzyme, ≤20 μL nuclease-free water, and 2.0 μL of each extracted chromosomal DNA. The amplification reaction for stx2 was carried out in 30 repeated temperature cycles in a thermal cycler block (ATC 401; Nyx Technik). The amplification reaction for stx2 was carried out in 30 repeated temperature changes called cycles each at 94°C for 2 minutes, denaturation at 94°C for 30 seconds, annealing at 56°C for 30 seconds, and extension at 72°C for 60 seconds.26,28
PCR amplification for hlyA
The hlyA amplification was carried out in a final reaction volume of 20 μL using 10 μL 2× PCR Master Mix, containing 1.5 μL MgCl2 (25 mM), 0.5 μL deoxyribonucleotide triphosphates (10 mM; deoxyadenosine triphosphate, deoxyguanosine triphosphate, deoxycytidine triphosphate, deoxythymidine triphosphate), 1 μL hlyA (2.5 μM) forward primer (5′-ACGATGTGGTTTATTCTGGA-3′) and reverse primer (5′-CTTCACGTGACCATACATAT-3′), 1 μL hlyA (2.5 μM), 0.2 μL Taq polymerase enzyme, ≤20 μL nuclease-free water, and 2.0 μL of each extracted chromosomal DNA. The amplification reaction for hlyA was carried out in 30 repeated temperature cycles in a thermal cycler block (ATC 401; Nyx Technik): at a temperature of 94°C for 2 minutes, denaturation at 94°C for 30 seconds, annealing at 45°C for 30 seconds, and extension at 72°C for 60 seconds.25,27,28
Amplicon assay
The stx1, stx2, and hlyA amplicons were electrophoresed in 1.5% agarose agar containing ethidium bromide (10 mg/mL) and a 1 kb standard marker at a current of 100 V for 30 minutes. They were then viewed under ultraviolet photo-documentation to determine their weight.25,26
Statistical analysis
The significance (P<0.05) of prevalent toxigenic E. coli was determined using a Student’s t-test, with a 95% confidence interval. Comparison of the resistant rates at different proportions was performed using the chi-squared test and Fisher’s exact test. The correlation of MIC to the zone of inhibition was determined using the method of least squares, with the zone diameter as the independent variable (X axis) and MIC as the dependent variable (Y axis).
Results
Table 1 shows the age distribution of the subjects whose fecal samples were studied. Of the 202 subjects recruited for the study, 125 (61.9%) were female and 77 (38.1%) were male. The age of the subjects ranged from <10 years to >40 years. The majority of the subjects screened fell within the ≥40-year-old age group (41.6%). Sorbitol fermenting and sorbitol nonfermenting E. coli (ie, STEC isolates) were identified in 86.1% of subjects, while the remaining 13.9% had NSTEC isolates (P=0.001).
  | Table 1 Bio-data distribution of the subjects |
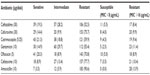
Table 2 shows the levels of resistance to antimicrobial agents among the 202 E. coli isolates analyzed by the disk diffusion test. The highest rate of resistance was against amoxicillin (90.6%), followed by 77.7% resistance to cefotaxime, 75.7% to cefuroxime, and 70.8% to ofloxacin. Twenty-one percent of the isolates were sensitive to cotrimoxazole and 20.3% to ofloxacin. In addition, 19.3% of the isolates were sensitive to ceftazidime, 14.9% to gentamicin, and 14.4% to cefuroxime. Low levels of sensitivity to antibiotics were only seen in cefotaxime (8.9%) and amoxicillin (3.5%).
To further validate the utility of the multiplex PCR assay, the NSTEC isolates of the 202 fecal samples were tested using a multiplex PCR where primers for only three virulence factors – stx1, stx2, and hlyA – were also investigated. Amplified DNA bands were as follows: stx2 (779 bp), stx1 (614 bp), and hlyA (165 bp) (data not shown). stx1 was the most commonly encountered factor, with its highest prevalence rate in the 0–10-year-old age group.
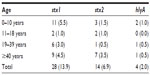
Table 3 shows the age distribution of sorbitol nonfermenting E. coli isolates harboring the Shiga toxin. The highest prevalence rate of stx1 (5.5%) was seen in the 0–10 year olds, with 4.5% in the ≥40 year olds. The highest prevalence rate of stx2 (3.5%) was seen in the ≥40 year olds, while the lowest prevalence rate of 0.5% was observed in the 19–39 year olds. A 1.0% prevalence rate of hlyA was observed among the 0–10 year olds and 0.5% in both the 19–39 and ≥40-year-old age groups. No prevalence of hlyA was recorded in the 11–18-year-old age group.
  | Table 3 Distribution of stx1, stx2, and hlyA in the different age groups |
Discussion
In this study, 41.6% of the subjects recruited for the study were aged ≥40 years and 17.8% were aged 0–10 years. Adults and young children are more prone to diarrhea or enteric infections in this locality. The young and elderly are at particular risk for progression of the disease to more severe sequelae; however, in healthy adults, infection may be restricted to watery diarrhea or may even be subclinical.29 A higher proportion of females (61.9%) and the 86.1% suspected STEC isolates obtained from the subjects indicate a steady rise in the spread of this serotype. This is lower than the 26% rate reported by Nielsen et al30 in a Danish study of 312 STEC infections, with 68% of the STEC isolates belonging to eight different pathogens. Although almost 500 pathogens of STEC have been isolated from humans with disease,31 it is associated with most cases of enteric diseases.
The introduction of antibiotics is one of the most important medical interventions with regard to reducing human morbidity and mortality. However, uncontrolled use of antibiotics has dramatically increased the frequency of resistance and prevalence of enteric infections, thereby reducing the therapeutic options available. Resistance dramatically reduces the possibility of treating infections effectively and increases the risk of complications and a fatal outcome. This is similar to the significantly high rate of STEC isolates resistant to amoxicillin (90.6%), cefotaxime (77.7%), and cefuroxime (75.7%). Resistance to amoxicillin, gentamicin, and cefotaxime was demonstrated with an MIC >16 μg/mL in 13.9%, 11.4%, and 10.4% of the isolates, respectively. The multidrug resistance found among these STEC isolates clearly presents a grave danger to the control of enteric infections caused by this virulent pathogen. High antimicrobial resistance observed in this study is similar to the E. coli isolates obtained from five geopolitical zones of Nigeria showing 94.4% resistance to ampicillin, 85.5% to cotrimoxazole, 92.1% to cefalotin, 78.7% to streptomycin, 70.8% to nitrofurantoin, 79.8% to tetracycline, 67.4% to chloramphenicol, 74.2% to amoxicillin, 61.8% to cefpirome, 52.8% to cefpodoxime, 46.1% to cefotaxime, 46.1% to ceftriaxone, 31.5% to cefoxitin, 38.2% to nalidixic acid, and 24.7% to gentamicin as reported by Nsofor et al.32 The patterns of resistance to commonly used antibiotics have been attributed in part to widespread and lengthy use of antibiotics.33 Constant exposure to antibiotic agents used for disease treatment, prophylaxis, or for livestock growth promotion necessitated plasmid mediation with a wide variety of genetic determinants.34 This makes it more possible for a susceptible bacterium to acquire resistance factors by conjugation or other genetic exchange mechanisms.
STEC is a known water-borne pathogen associated with diarrhea, HC, HUS, and renal failure in children.11 The prevalence rates of 13.9%, 6.9%, and 2.0% obtained for stx1, stx2, and hlyA, respectively, in this study is a threat to public health. The virulence rate of 5.5% for stx1 in 0–10 year olds and 3.5% for stx2 in ≥40 year olds demands urgent attention.
The marked heterogeneity of the STEC strains identified in this study showed that the spread of STEC seems much more complex and it could keep spreading from animals to humans and from person to person. The prevalence rates reported here are lower than those for stx1 (56%) and stx2 (43%) in STEC isolates reported by Yakubu et al.19 These are markedly high and the presence of either or both has been related to the ability of STEC strains to cause serious disease in humans. Only stx2 has been shown to be more commonly associated with serious human diseases compared with strains producing stx1 alone or both stx1 and stx2, possibly because the level of transcription of stx2 in vivo is higher than that of stx1.13 The production of the Shiga toxin by STEC is the primary virulence trait responsible for HUS. In addition, STEC strains are the major cause of gastroenteritis and HC, and are also associated with renal failure in children.35 However, a significant minority of human STEC isolates, including those from patients with HC and HUS, produce enterohemolysin (hlyA product) which might have a direct association with the capacity of a given STEC strain to cause more serious disease.36
The multiplex PCR described in this study is a better way of detecting the virulence of these genes in humans. A number of primers which amplify these genes have previously been reported, including the ones used in this study.27,28 The combination of primers reported here was chosen to generate a high level of sensitivity to the tested isolates. In this study, none of the isolates carried more than one gene from the sampled isolates. The distribution of these genes is a reflection of the prevalence ratio in relation to the studied age groups. Other report shows that patterns of STEC are affected by diet, age, stress, and seasonal variation.37
Table 4 shows the various primers used in this study. As previously stated, the presence of these genes is obvious in this environment. In addition, it is well known that STEC is a zoonotic food and water-borne pathogen associated with diarrhea, HC, HUS, and renal failure, particularly in children, and can cause attaching and effacing properties in diarrhea cases.2,11,12 The findings of these reports justify their presence and the need for further testing and characterization of STEC and NSTEC in this zone and other parts of Nigeria.
  | Table 4 List of primer sequences used |
It is obvious from this study that the prevalence of virulent STEC infections is a public health concern in Nigeria, and the use of a relevant molecular tool such as a PCR assay should aid quick detection of this virulent serotype and help to curb severe epidemic human diseases associated with STEC infections. Based on this study, it could be suggested that the common incidence of renal failure in children and adolescents in this locality may be a result of virulent antibiotic-resistant STEC through consumption of contaminated meat (especially undercooked beef hamburgers), ready-to-eat sausages, raw milk, cheese, unpasteurized milk, and drinking water as well as contact with animals.18 The overall prevalence rates of these genes shows a need to step up control measures to curb the spread of STEC in this environment.
However, it has been reported that the potential harmful or beneficial effects of antibiotic treatment may be as dependent on the properties of resident bacterial flora as on the infecting O157:H7 strain.38 The importance of antibiotic resistance in STEC-infected individuals cannot be overemphasized as it further complicates the disease condition through the release of toxins from the Gram-negative wall of the bacterium. It further clarifies that the role of antibiotics for treatment of E. coli O157:H7 infections is controversial and complex. The high prevalence of antimicrobial resistance observed in all of the isolates in this study, especially to amoxicillin, cefotaxime, and cefuroxime, further confirms that the release of the Shiga toxin might not be unrelated to the presence of resistance. It is an interesting example of a situation in which antibiotic treatment can harm the patient,38,39 and shows that administration of antibiotics can worsen the condition of a STEC-infected individual.
Conclusion
Poor detection of HUS and severe colitis caused by misdiagnosed STEC infection has greatly contributed to the prevalence of STEC infection and increased morbidity. Therefore, use of a PCR assay for STEC detection is very precise and effective for surveillance of HUS and other renal infections.
Acknowledgments
The management and staff of the study centers are hereby acknowledged; in particular, members of the Department of Biotechnology, Federal University of Agriculture (Abeokuta, Nigeria) and the Department of Medical Microbiology, Ladoke Akintola University of Technology (Osogbo Campus, Nigeria).
Disclosure
The authors report no conflicts of interest in this work.
References
Gyles CL. Escherichia coli cytotoxins and enterotoxins. Can J Microbiol. 1992;38(7):734–746. | |
Clarke SC, Haigh RD, Freestone PP, Williams PH. Virulence of enteropathogenic Escherichia coli, a global pathogen. Clin Microbiol Rev. 2003;16(3):365–378. | |
Blanco J, Blanco M, Blanco JE, et al. Verotoxin-producing Escherichia coli in Spain: prevalence, serotypes, and virulence genes of O157:H7 and non-O157 VTEC in ruminants, raw beef products, and humans. Exp Biol Med (Maywood). 2003;228(4):345–351. | |
Griffin PM, Tauxe RV. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol Rev. 1991;13:60–98. | |
World Health Organization. Manual for Laboratory Investigation of Acute Enteric Infections. Geneva: World Health Organization; 1987. | |
Browning NG, Botha JR, Sacho H, Moore PJ. Escherichia coli O157 : H7 haemorrhagic colitis. Report of the first South African case. S Afr J Surg. 1990;28(1):28–29. | |
Donnenberg MS, Kaper JB. Enteropathogenic Escherichia coli. Infect Immun. 1992;60(10):3953–3961. | |
Frankel G, Phillips AD, Rosenshine I, Dougan G, Kaper JB, Knutton S. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol Microbiol. 1998;30(5):911–921. | |
Al-Gallas N, Ben Aissa R, Annabi AT, Bahri O, Boudabous A. Isolation and characterization of Shiga-toxin producing Escherichia coli from meat and dairy products. Food Microbiol. 2002;19(4):389–398. | |
Olanipekun OO. Prevalence of Enteropathogenic Escherichia Coli in Children with Diarrhoea Attending Jos University Teaching Hospital [MSc Thesis]. Jos: University of Jos; 1996. | |
Coombes KB, Wickham ME, Mascarenhas M, Gruenheid S, Finlay BB, Karmali MA. Molecular analysis as an aid to assess the public health risk of non-O157 Shiga toxin-producing Escherichia coli strains. Appl Environ Microbiol. 2008;74(1):2153–2160. | |
Nunes BE, Saridakis OH, Irino K, Pelayo JS. Genotypic and phenotypic characterization of attaching and effacing Escherichia coli (AEEC) which were isolated from children with and without diarrhoea in Londrina, Brazil. J Med Microbiol. 2003;52(Pt 6):499–504. | |
Sung LM, Jackson MP, O’Brien AD, Holmes RK. Transcription of the Shiga-like toxin type II and Shiga-like toxin type II variant operons of Escherichia coli. J Bacteriol. 1990;172(11):6386–6395. | |
Barman NN, Deb R, Ramamurthy T, et al. Molecular characterization of Shiga-like toxin-producing Escherichia coli (STEC) isolates from pigs oedema. Indian J Med Res. 2008;127(6):602–606. | |
Bennett J, Bettelheim KA. Serotypes of non-O157 verocytotoxigenic Escherichia coli isolated from meat in New Zealand. Comp Immunol Microbiol Infect Dis. 2002;25(2):77–84. | |
Meichtri L, Miliwebsky E, Gioffre A, et al. Shiga toxin-producing Escherichia coli in healthy young beef steers from Argentina: prevalence and virulence properties. Int J Food Microbiol. 2004;96(2):189–198. | |
Schmidt H, Karch H. Enterohemolytic phenotypes and genotypes of Shiga-toxin-producing Escherichia coli O111 strains from patients with diarrhea and hemolytic-uremic syndrome. J Clin Microbiol. 1996;34(10):2364–2367. | |
Karmali MA, Steele BT, Petric M, Lim C. Sporadic cases of haemolytic-uraemic syndrome associated with faecal cytotoxin and cytotoxin-producing Escherichia coli in stools. Lancet. 1983;1(8325):619–620. | |
Beutin L, Miko A, Krause G, et al. Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by a combination of serotyping and molecular typing of Shiga toxin genes. Appl Environ Microbiol. 2007;73(15):4769–4775. | |
Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11(1):142–201. | |
Yakubu B, Boniface ND, Umaru DA. Public health risk of O157 and non-O157 Shiga toxin-producing Escherichia coli strains in a semi-urban environment in Nigeria. Afr J Microbiol Res. 2011;5(31):5636–5643. | |
National Population Commission. National Census Sentinell, Nigeria. 2007. | |
Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45(4):493–496. | |
Clinical and Laboratory Standards Institute. M100-S19: Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute; 2009. | |
Minas K, McEwan NR, Newbold CJ, Scott KP. Optimization of a high-throughput CTAB-based protocol for the extraction of qPCR-grade DNA from rumen fluid, plant and bacterial pure cultures. FEMS Microbiol Lett. 2011;325(2):162–169. | |
Willner D, Daly J, Whiley D, Grimwood K, Wainwright CE, Hugenholtz P. Comparison of DNA extraction methods for microbial community profiling with an application to pediatric broncholalveolar lavage samples. PLoS One. 2012;7(4):e34605. | |
Fratamico PM, Sackitey SK, Wiedmann M, Deng MY. Detection of Escherichia coli O157:H7 by multiplex PCR. J Clin Microbiol. 1995;33(8):2188–2191. | |
Gannon VP, King RK, Kim JY, Golsteyn Thomas EJ. Rapid and sensitive method for detection of Shiga-like toxin-producing Escherichia coliin ground beef using the polymerase chain reaction. Appl Environ Microbiol. 1992;58(12):3809–3815. | |
Smith-Palmer A, Locking M, Reilly B, Fisher I. Cluster of E. coli O157 infections in Scottish tourists returning from southwest Turkey, Jul–Aug 2005. Euro Surveill. 2005;10(8):E050818.2. | |
Nielsen EM, Scheutz F, Torpdahl M. Continuous surveillance of Shiga toxin-producing Escherichia coli infections by pulsed-field gel electrophoresis shows that most infections are sporadic. Foodborne Pathog Dis. 2006;3(1):81–87. | |
Blanco M, Schumache S, Tasara T, et al. Serotypes, intimin variants, and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-eta2). BMC Microbiol. 2005;5:23. | |
Nsofor CA, Iroegbu CU. Antibiotic resistance profile of Escherichia coli isolated from five major geopolitical zones of Nigeria. African Journal of Bacteriology Research. 2013;5(3):29–34. | |
Piddock LJ. Multidrug-resistance efflux pumps – not just for resistance. Nat Rev Microbiol. 2006;4(8):629–636. | |
van den Bogaard AE, London N, Driessen C, Stobberingh EE. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers, and poultry slaughterers. J Antimicrob Chemother. 2001;47(6):763–771. | |
Todd WT, Dundas S. The management of VTEC O157 infection. Int J Food Microbiol. 2001;66(1–2):103–110. | |
Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36(2):598–602. | |
Kudva IT, Hatfield PG, Hovde CJ. Escherichia coli O157:H7 in microbial flora of sheep. J Clin Microbiol. 1996;34(2):431–433. | |
Craig-Mylius KA, Weiss AA. Antibacterial agents and release of periplasmic pertussis toxin from Bordetella pertussis. Antimicrob Agents Chemother. 2000;44(5):1383–1386. | |
Grif K, Dierich MP, Karch H, Allerberger F. Strain-specific differences in the amount of Shiga toxin released from enterohemorrhagic Escherichia coli O157 following exposure to subinhibitory concentrations of antimicrobial agents. Eur J Clin Microbiol Infect Dis. 1998;17(11):761–766. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.