Back to Journals » Neuropsychiatric Disease and Treatment » Volume 10
Genetic association between the dopamine D1-receptor gene and paranoid schizophrenia in a northern Han Chinese population
Authors Yao J , Ding M, Xing J, Xuan J, Pang H, Pan Y, Wang B
Received 23 January 2014
Accepted for publication 20 February 2014
Published 17 April 2014 Volume 2014:10 Pages 645—652
DOI https://doi.org/10.2147/NDT.S61227
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Jun Yao, Mei Ding, Jiaxin Xing, Jinfeng Xuan, Hao Pang, Yuqing Pan, Baojie Wang
Institute of Forensic Medicine, China Medical University, Shenyang, People's Republic of China
Objective: Dysregulation of dopaminergic neurotransmission at the D1 receptor in the prefrontal cortex has been implicated in the pathogenesis of schizophrenia. Genetic polymorphisms of the dopamine D1-receptor gene have a plausible role in modulating the risk of schizophrenia. To determine the role of DRD1 genetic polymorphisms as a risk factor for schizophrenia, we undertook a case-control study to look for an association between the DRD1 gene and schizophrenia.
Materials and methods: We genotyped eleven single-nucleotide polymorphisms within the DRD1 gene by deoxyribonucleic acid sequencing involving 173 paranoid schizophrenia patients and 213 unrelated healthy individuals. Statistical analysis was performed to identify the difference of genotype, allele, or haplotype distribution between cases and controls.
Results: A significantly lower risk of paranoid schizophrenia was associated with the AG + GG genotype of rs5326 and the AG + GG genotype of rs4532 compared to the AA genotype and the AA genotype, respectively. Distribution of haplotypes was no different between controls and paranoid schizophrenia patients. In the males, the genotype distribution of rs5326 was statistically different between cases and controls. In the females, the genotype distribution of rs4532 was statistically different between cases and controls. However, the aforementioned statistical significances were lost after Bonferroni correction.
Conclusion: It is unlikely that DRD1 accounts for a substantial proportion of the genetic risk for schizophrenia. As an important dopaminergic gene, DRD1 may contribute to schizophrenia by interacting with other genes, and further relevant studies are warranted.
Keywords: dopamine D1 receptor, paranoid schizophrenic, single-nucleotide study, association, genetic polymorphism
Introduction
Schizophrenia is a common mental disorder effected by the mutual influence of multiple genetic1 and environmental factors,2 and its heritability is up to 80%.3 In order to clarify the pathological mechanisms, many studies have been carried out over the years, but a consistent conclusion has still not been achieved. To date, dysregulation of dopaminergic neurotransmission has been implicated in the pathogenesis of schizophrenia.4,5 The reduction of prefrontal cortical dopamine neurotransmission likely leads to schizophrenia.6
Dopamine is an important endogenous neurotransmitter that plays a significant role in modulating cognitive, mood, and motor functions of the brain.7 It plays a regulatory function by binding to the dopamine receptors of the postsynaptic membrane. The sequence of the dopamine D1 receptor gene (DRD1) has been identified and mapped to chromosome 5q35.1.8 Linkage between DRD1 and schizophrenia has been reported in Chinese,9 American,10 and Portuguese populations.11 As we all know, a remarkable symptom of schizophrenia patients is cognitive dysfunction (such as attention-deficit and memory disorders).6,12 Interestingly, an optimal amount of DRD1 stimulation is essential in maintaining normal prefrontal cognitive function.13,14 Studies15–17 show that deficits in DRD1 transmission are associated with cognitive and negative symptoms of schizophrenia, and there is a significant decrease in DRD1 expression in the basal ganglia of schizophrenic patients. DRD1 gene polymorphisms likely play a role in the occurrence of schizophrenia by affecting the expression of the D1 receptor. The rs1799914 genetic polymorphism is associated with schizophrenia in a Korean population.18 A study of five single-nucleotide polymorphisms (SNPs; rs11746641, rs11749676, rs251937, rs12518222, rs4867798) has reported that rs11746641 and rs11749676 are associated with schizophrenia in males, and the haplotype T-A-T-C-T can reduce the risk of schizophrenia.19 The genetic polymorphism of rs4532 is likely associated with deficits in executive function and performance on the Wisconsin Card Sorting Test.6 However, no association between rs4532 and schizophrenia has been reported,20,21 and a consistent conclusion has still not been achieved.
Many previous studies analyzed a few loci (one to five SNPs) or selected several tag SNPs as a substitute for the entire gene region. However, the limitation of several loci could not completely pinpoint the true susceptible SNPs, owing to the weak linkage disequilibrium in the DRD1 gene. Most likely, the basis of inconsistent results was due to this factor. We analyzed eleven SNPs in the 5′-flanking and untranslated regions of the DRD1 gene by deoxyribonucleic acid (DNA) sequencing, and carried out a case-control study between 173 paranoid schizophrenia patients and 213 unrelated healthy controls.
Materials and methods
Samples
Blood samples from 213 healthy, unrelated, northern Han Chinese volunteers (112 males and 101 females, average age 40.1±14.7 years) were provided by China Medical University’s Department of Blood Serum in the School of Forensic Medicine. Questionnaires confirmed there was no history of mental disease going back three generations. Blood samples from 173 northern Han Chinese patients with paranoid schizophrenia (88 males and 85 females, average age 43.7±13.5 years) were provided by the Third People’s Hospital of Liaoning Province. Each patient was diagnosed by at least two senior psychiatrists. Diagnostic criteria for paranoid schizophrenia were in line with the Diagnostic and Statistical Manual of Mental Disorders (fourth edition)22 on the basis of unstructured interviews and information from medical records. The study protocol and process was assessed and approved by the ethics committee at China Medical University. In the present study, all subjects signed the informed consent form.
Selection of polymorphic loci
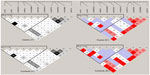
Polymerase chain reaction (PCR) was used to amplify DRD1 fragment length, including 5′-flanking and untranslated regions. The nucleotide position of one fragment amplified was from –3,052 to –1,483 (primer sequence [forward] – ctgatatggtgcatggctgtt, [reverse] – acctgcgttgtctccaagtgt). The nucleotide position of the other fragment amplified was from –1,505 to +100 (primer sequence [forward] – ggacacttggagacaacgcag, [reverse] – atgagcagcgacaggaaacag). The amplified fragment included 45 SNP loci that had been reported in the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/gene/),39 and were genotyped by DNA sequencing. The frequencies of the SNP loci (>1%) were screened. Eleven polymorphic loci were detected in the northern Han Chinese population, including one newly discovered SNP loci: ss492961114/rs201089398 in the 5′ regulatory region. A schematic diagram for DRD1 is shown in Figure 1.
Genotyping
Genomic DNA was extracted by the traditional phenol-chloroform method and quantified with ultraviolet (UV) spectrophotometry. An ABI9700 amplifier (Applied Biosystems, Foster City, CA, USA) was used to amplify the target band. Beijing Genomics Institute (ABI3730XL) was commissioned for sequencing.
Statistical analysis
SPSS 18.0 software (IBM, Armonk, NY, USA) was used to calculate genotype frequency and allele frequency, and Haploview 4.1 software (Broad Institute, Cambridge, MA, USA) was used for the Hardy–Weinberg equilibrium test, the confirmation of haplotypes, and analysis of intergroup differences. The χ2 test was used to measure the association of paranoid schizophrenic risk with DRD1 genotypes, alleles, and haplotypes between control and disease groups. Unconditional logistic regression models were used to obtain maximum-likelihood estimates of odds ratios (ORs) and their 95% confidence intervals (CIs) between each locus and the presence of paranoid schizophrenia. The Bonferroni correction was used in multiple testing, and P-values were divided by the total number of loci or haplotypes.
Results
Haploview 4.1 was used to test genotype fitness for the control and paranoid schizophrenic groups. The results showed that the distribution of no genotype frequencies at all loci deviated from the Hardy–Weinberg equilibrium in either group or in the male and female subgroups (P>0.05). Results of linkage analysis for control and disease groups are shown in Figure 2.
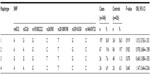
Genotype and allele frequencies of eleven SNP loci in DRD1 are shown in Table 1. The χ2 test was used to measure the association of paranoid schizophrenic risk with DRD1 genotypes. Among them, the genotype distribution of rs5326 was statistically different between paranoid schizophrenia patients and controls (P=0.036), though the difference was lost after Bonferroni correction. Compared with the AA genotype, the AG + GG genotype reduced the disease risk (ORAG + GG 0.358, 95% CIAG + GG 0.151–0.851). In addition, a lower risk of paranoid schizophrenia was associated with the AG + GG genotype of rs4532 (ORAG + GG 0.592, 95% CIAG + GG 0.350–1.001) compared with the AA genotype. No remaining loci exhibited significant differences in allele or genotype frequencies between the patients and controls.
A comparison of the frequencies of haplotypes formed by the seven SNPs in the block was analyzed between the patients and controls. The results are shown in Table 2. The remaining four SNPs outside the block formed the haplotypes, and the result is shown in Table 3. All-haplotype analysis found that there was no association with the occurrence of paranoid schizophrenia between patients and controls.
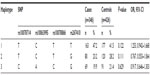
A study reported that sexually dimorphic DRD1 SNPs impacted on the risk of schizophrenia.19 Therefore, we stratified the observed association of rs5326 and rs4532 with the risk of paranoid schizophrenia based on sex by the χ2 test (results shown in Table 4). In males, the genotype distribution of rs5326 was statistically different between cases and controls (P=0.048). In females, the genotype distribution of rs4532 was statistically different between cases and controls (P=0.029). However, these significant differences disappeared after Bonferroni correction.
Discussion
In the present study, we investigated eleven SNPs in the DRD1 gene in a northern Chinese Han population by DNA sequencing. There were no significant differences in allele, genotype, or haplotype frequencies between paranoid schizophrenia patients and controls after Bonferroni correction. Consistent with our results, some studies reported no significant association between the DRD1 gene and schizophrenia.20,23 However, several previous studies have reported that the DRD1 gene is associated with bipolar disorder,21 attention deficit with hyperactivity disorders,24 nicotine dependence,25 alcohol dependence,26 and others. Compared with these diseases, schizophrenic patients have the same or similar symptoms.
Our results found that the AG + GG genotype of rs5326, as a protective factor for schizophrenia, could reduce the risk of paranoid schizophrenia, as assessed by the χ2 test. Zhu et al27 reported that rs5326 was not associated with schizophrenia in the Chinese Han population, which was consistent with previous studies.28,29 The interaction between the DNMT3B and DRD1 genes significantly increased the risk of schizophrenia after analysis with multifactor dimensionality reduction software.29 This also shows that the incidence of schizophrenia is affected by the interaction of multiple genes, and studies of only one gene cannot clearly explain the pathogenesis. Although some studies20,26,30 found no association between rs4532 and schizophrenia, a meta-analysis31 indicated that DRD1 might contain a genuine susceptibility allele, and the OR of rs4532 reached 1.18 (95% CI 1.01–1.38). In addition, rs4532 was associated with antipsychotic treatment in schizophrenia32,33 and working memory and cognitive behavior related to the prefrontal cortex.6,13 This shows that rs4532 likely has a potential link with the etiology of schizophrenia. Furthermore, the inconsistency of association studied between DRD1 and schizophrenia may result from genetic heterogeneity, which is an important challenge in the genetic study of schizophrenia. In our study, we reported rs201089398 for the first time. However, this new locus had a slightly lower frequency, and we failed to reveal its potential association with schizophrenia in the Chinese Han population.
The pathogenesis of schizophrenia is the participation of a variety of risk factors, and sex is an important factor.34,35 Our study showed that rs5326 was statistically different between cases and controls in males and rs4532 was statistically different between cases and controls in females by the χ2 test. Unfortunately, the impact of sex-specific mechanisms in schizophrenia is unclear, and needs further exploration in the future. In addition, there was another limitation in our study in that we did not analyze positive and negative symptoms or cognitive performance of the patients, owing to the lack of original information.
What can we learn from these results? First, the analysis of just one gene is far from enough, because DRD1 may result in schizophrenia by interacting with other dopaminergic-related genes, such as synthase genes, metabolic genes, transporter genes, and others. Additionally, other neurotransmitters are also involved in the pathogenesis of schizophrenia. We cannot definitively rule out a role for any of these genes in schizophrenia. Therefore, further studies should be focused on genome-wide SNP analysis and gene–gene interactions to determine their possible roles in the etiology of schizophrenia.
Second, multiethnic studies with large samples are essential. Many of the ORs for association are in a plausible range (1.10–1.23) for small susceptibility effects, but below what would produce significant P-values in smaller samples.36 The larger ORs in some previous reports may either be false positives or inflated estimates of genetic effects, while association studies of large samples provide more power. In the present study, we reported rs201089398 for the first time. With regard to the limited samples, the slightly lower frequency of this new locus might not reveal the potential association with schizophrenia in the Chinese Han population. Besides, our results demonstrated that the genotype frequencies of DRD1 are variable in different ethnic groups. Therefore, through multiethnic studies, it will likely be easier to find a genetic effect for susceptibility to schizophrenia.
Third, genetic heterogeneity is a significant challenge in the study of schizophrenia,37 and family-based studies could overcome this problem effectively.38 Our results need to be clarified through analysis of extensive family studies. Additionally, paranoid schizophrenia, as a subtype of schizophrenia, includes positive and negative symptoms, which show different clinical manifestations. Unfortunately, due to the limited data, we did not do a more detailed analysis. Overall, the shortcomings of our study could partially inform future researchers.
In summary, although our study did not find a genetic association between DRD1 and paranoid schizophrenia, its possible role in the risk of schizophrenia cannot be definitively excluded. Further and larger studies are necessary to evaluate the effect of DRD1 polymorphisms on the etiology of schizophrenia, and our data may provide a reference.
Acknowledgment
Special thanks to the Third People’s Hospital of Liaoning Province for providing blood samples from paranoid schizophrenic patients for this experiment.
Disclosure
The authors report no conflicts of interest in this work.
References
Gill P, Gusmão L, Haned H, et al. DNA commission of the International Society of Forensic Genetics: recommendations on the evaluation of STR typing results that may include drop-out and/or drop-in using probabilistic methods. Forensic Sci Int Genet. 2012;6:679–688. | |
Mjellem N, Kringlen E. Schizophrenia: a review, with emphasis on the neurodevelopmental hypothesis. Nord J Psychiatry. 2001;55:301–309. | |
Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. | |
Howes OD, Kapur S. The dopamine hypothesis of schizophrenia: version III – the final common pathway. Schizophr Bull. 2009;35:549–562. | |
Abi-Dargham A, Moore H. Prefrontal DA transmission at D1 receptors and the pathology of schizophrenia. Neuroscientist. 2003;9:404–416. | |
Rybakowski JK, Borkowska A, Czerski PM, Kapelski P, Dmitrzak-Weglarz M, Hauser J. An association study of dopamine receptors polymorphisms and the Wisconsin Card Sorting Test in schizophrenia. J Neural Transm. 2005;112:1575–1582. | |
Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine receptors: from structure to function. Physiol Rev. 1998;78:189–225. | |
Grandy DK, Zhou QY, Allen L, et al. A human D1 dopamine receptor gene is located on chromosome 5 at q35.1 and identifies an EcoRI RFLP. Am J Hum Genet. 1990;47:828–834. | |
Zheng Y, Wang X, Gu N, et al. A two-stage linkage analysis of Chinese schizophrenia pedigrees in 10 target chromosomes. Biochem Biophys Res Commun. 2006;342:1049–1057. | |
Escamilla MA, Ontiveros A, Nicolini H, et al. A genome-wide scan for schizophrenia and psychosis susceptibility loci in families of Mexican and Central American ancestry. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:193–199. | |
Sklar P, Pato MT, Kirby A, et al. Genome-wide scan in Portuguese Island families identifies 5q31-5q35 as a susceptibility locus for schizophrenia and psychosis. Mol Psychiatry. 2004;9:213–218. | |
Rinaldi A, Mandillo S, Oliverio A, Mele A. D1 and D2 receptor antagonist injections in the prefrontal cortex selectively impair spatial learning in mice. Neuropsychopharmacology. 2007;32:309–319. | |
Williams GV, Castner SA. Under the curve: critical issues for elucidating D1 receptor function in working memory. Neuroscience. 2006;139:263–276. | |
Meyer-Lindenberg A, Kohn PD, Kolachana B, et al. Midbrain dopamine and prefrontal function in humans: interaction and modulation by COMT genotype. Nat Neurosci. 2005;8:594–596. | |
Okubo Y, Suhara T, Suzuki K, et al. Decreased prefrontal dopamine D1 receptors in schizophrenia revealed by PET. Nature. 1997; 385:634–636. | |
Karlsson P, Farde L, Halldin C, Sedvall G. PET study of D(1) dopamine receptor binding in neuroleptic-naive patients with schizophrenia. Am J Psychiatry. 2002;159:761–767. | |
Abi-Dargham A, Mawlawi O, Lombardo I, et al. Prefrontal dopamine D1 receptors and working memory in schizophrenia. J Neurosci. 2002;22:3708–3719. | |
Lee KY, Joo EJ, Ji YI, et al. Associations between DRDs and schizophrenia in a Korean population: multi-stage association analyses. Exp Mol Med. 2011;43:44–52. | |
Hoenicka J, Garrido E, Ponce G, et al. Sexually dimorphic interaction between the DRD1 and COMT genes in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:948–954. | |
Zhang C, Fang Y, Xie B, et al. No genetic association between dopamine D1 receptor gene and [early onset] schizophrenia. Psychiatry Res. 2010;177:350–353. | |
Dmitrzak-Weglarz M, Rybakowski JK, Slopien A, et al. Dopamine receptor D1 gene – 48A/G polymorphism is associated with bipolar illness but not with schizophrenia in a Polish population. Neuropsychobiology. 2006;53:46–50. | |
American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders (4de herz. versie). Washington: American Psychiatric Association; 2000. | |
Hoogendoorn ML, Bakker SC, Schnack HG, et al. No association between 12 dopaminergic genes and schizophrenia in a large Dutch sample. Am J Med Genet B Neuropsychiatr Genet. 2005; 134B:6–9. | |
Bobb AJ, Addington AM, Sidransky E, et al. Support for association between ADHD and two candidate genes: NET1 and DRD1. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:67–72. | |
Huang W, Ma JZ, Payne TJ, Beuten J, Dupont RT, Li MD. Significant association of DRD1 with nicotine dependence. Hum Genet. 2008; 123:133–140. | |
Batel P, Houchi H, Daoust M, Ramoz N, Naassila M, Gorwood P. A haplotype of the DRD1 gene is associated with alcohol dependence. Alcohol Clin Exp Res. 2008;32:567–572. | |
Zhu F, Yan CX, Wang Q, et al. An association study between dopamine D1 receptor gene polymorphisms and the risk of schizophrenia. Brain Res. 2011;1420:106–113. | |
Zhang C, Wu ZG, Shao Y, Li ZZ, Yu SY, Fang YR. [Association analysis between dopamine D1 receptor gene and symptom quantitative trait of schizophrenia]. Zhonghua Yi Xue Za Zhi. 2011;91:2019–2022. Chinese. | |
Zhang C, Xie B, Du YS, et al. [Gene-gene interaction between DNMT3B and DRD1 in schizophrenia]. Zhonghua Yi Xue Za Zhi. 2010;90:3059–3062. | |
Kojima H, Ohmori O, Shinkai T, Terao T, Suzuki T, Abe K. Dopamine D1 receptor gene polymorphism and schizophrenia in Japan. Am J Med Genet. 1999;88:116–119. | |
Allen NC, Bagade S, McQueen MB, et al. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nat Genet. 2008;40:827–834. | |
Potkin SG, Basile VS, Jin Y, et al. D1 receptor alleles predict PET metabolic correlates of clinical response to clozapine. Mol Psychiatry. 2003;8:109–113. | |
Hwang R, Shinkai T, De Luca V, et al. Association study of four dopamine D1 receptor gene polymorphisms and clozapine treatment response. J Psychopharmacol. 2007;21:718–727. | |
Canuso CM, Pandina G. Gender and schizophrenia. Psychopharmacol Bull. 2007;40:178–190. | |
Pregelj P. Neurobiological aspects of psychosis and gender. Psychiatr Danub. 2009;21 Suppl 1:128–131. | |
Sanders AR, Duan J, Levinson DF, et al. No significant association of 14 candidate genes with schizophrenia in a large European ancestry sample: implications for psychiatric genetics. Am J Psychiatry. 2008; 165:497–506. | |
Meng J, Shi Y, Zhao X, et al. No significant association between the genetic polymorphisms in the GSK-3 beta gene and schizophrenia in the Chinese population. J Psychiatr Res. 2008;42:365–370. | |
Won S, Lange C. A general framework for robust and efficient association analysis in family-based designs: quantitative and dichotomous phenotypes. Stat Med. Epub June 6, 2013. | |
Benson DA, Cavanaugh M, Clark K, et al. GenBank. Nucleic Acids Res. 2013;41:D36–42. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.