Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Efficient gene delivery system mediated by cis-aconitate-modified chitosan-g-stearic acid micelles
Authors Yao J, Du Y, Yuan H , You J, Hu F
Received 21 January 2014
Accepted for publication 19 February 2014
Published 18 June 2014 Volume 2014:9(1) Pages 2993—3003
DOI https://doi.org/10.2147/IJN.S61103
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Jing-Jing Yao, Yong-Zhong Du, Hong Yuan, Jian You, Fu-Qiang Hu
College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, People’s Republic of China
Abstract: Cis-aconitate-modified chitosan-g-stearic acid (CA-CSO-SA) micelles were synthesized in this study to improve the gene transfection efficiency of chitosan-g-stearic acid (CSO-SA). The CA-CSO-SA micelles had a similar size, critical micelle concentration, and morphology, but their zeta potential and cytotoxicity were reduced compared with CSO-SA micelles. After modification with cis-aconitate, the CA-CSO-SA micelles could also compact plasmid DNA (pDNA) to form nanocomplexes. However, the DNA binding ability of CA-CSO-SA was slightly reduced compared with that of CSO-SA. The transfection efficiency mediated by CA-CSO-SA/pDNA against HEK-293 cells reached up to 37%, and was much higher than that of CSO-SA/pDNA (16%). Although the cis-aconitate modification reduced cellular uptake kinetics in the initial stages, the total amount of cellular uptake tended to be the same after 24 hours of incubation. An endocytosis inhibition experiment showed that the internalization mechanism of CA-CSO-SA/pDNA in HEK-293 cells was mainly via clathrin-mediated endocytosis, as well as caveolae-mediated endocytosis and macropinocytosis. Observation of intracellular trafficking indicated that the CSO-SA/pDNA complexes were trapped in endolysosomes, but CA-CSO-SA/pDNA was more widely distributed in the cytosol. This study suggests that modification with cis-aconitate improves the transfection efficiency of CSO-SA/pDNA.
Keywords: chitosan-g-stearic acid, cis-aconitate, micelles, transfection efficiency, intracellular trafficking
Introduction
Nucleic acid-based therapeutics are envisaged to play a significant role in the next generation of treatments for a variety of diseases, such as cancer.1,2 Because nucleic acids are susceptible to hydrolytic and enzymatic degradation in the body and cannot pass through the cytoplasmic membrane, a safe and efficient delivery vehicle is necessary to enable nucleic acids to realize their maximum therapeutic potential under harsh in vivo conditions.3 In recent years, polysaccharides and their derivatives have been widely investigated for their potential application as carriers for gene delivery.4,5 An example is chitosan, which has several attractive properties, including biocompatibility, biodegradability, and availability. However, chitosan has some drawbacks in gene delivery systems, in particular inefficient endosomal escape, failure of DNA dissociation, and low transfection efficiency,6 and many researchers have tried to overcome these problems.7–12 One approach taken to improving transport from the endosomal compartment to the cytoplasm has been to incorporate fusogenic viral peptides and polyacrylic acid in which the protonated carboxyl group plays an important role.13,14 Some researchers have also demonstrated that addition of negatively charged polyanions can improve the in vitro transfection efficiency of chitosan and other polycations by loosening the polymer/DNA complexes.15–17
In a previous study, we modified chitosan using hydrophobic stearic acid (SA) to form self-assembling micelles. It has been demonstrated that chitosan oligosaccharide (CSO)-SA micelles are not only a promising vehicle for antitumor drug delivery,18,19 but are also effective gene carriers.20,21 CSO-SA can self-aggregate to form micelles in aqueous solution and condense DNA into nanocomplexes easily, with excellent internalization into cancer cells.22 However, CSO-SA/plasmid DNA (pDNA) nanocomplexes may also have problems with regard to endolysosomal escape and DNA dissociation, resulting in unsatisfactory transfection efficiency. According to our previous study, polyethylenimine-conjugated CSO-SA can suppress tumor growth effectively (by more than 60%) using plasmid pigment epithelium-derived factor as a therapeutic gene.23 The effectiveness of this antitumor gene therapy may be ascribed at least in part to the proton-buffering ability of polyethylenimine that could promote endolysosomal escape. Incorporation of bovine serum albumin with an isoelectric point of 4.7 into CSO-SA/pDNA resulted in better transfection efficiency than that found with CSO-SA/pDNA complexes alone, while incorporation of bovine serum albumin with an isoelectric point of 9.3 was unfavorable for the transfection efficiency of CSO-SA/pDNA complexes. These findings indicate that a degree of negative charge or an acidic group is beneficial for gene transfection using CSO-SA/pDNA.24
Cis-aconitate (CA) has three carboxyl residues and, being an intermediate of the tricarboxylic acid cycle, is an endogenous biological agent. We hypothesized that modification of CSO-SA with CA could be one of the solutions to enhancing the gene transfection efficiency of CSO-SA/pDNA. In the present study, we grafted CA onto CSO-SA via amide bonding, and evaluated the resulting CA-CSO-SA micelles for their size, zeta potential, pDNA condensation ability, and DNA protection ability. Their gene transfection ability was investigated in HEK-293 cells using pDNA encoding enhanced green fluorescent protein (pEGFP-C1) as the reporter gene. Cellular uptake and intracellular trafficking were observed by confocal laser scanning microscopy. The endocytosis pathway was also investigated using endocytic inhibitors.
Materials and methods
Materials
CSO with an average molecular weight of about 15.0 kDa was obtained by enzymatic degradation of 95% deacetylated chitosan (molecular weight 45.0 kDa) supplied by Yuhuan Marine Biochemistry Co Ltd (Zhejiang, People’s Republic of China). The SA was from Shanghai Chemical Reagent Co Ltd (Shanghai, People’s Republic of China), and 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC), and 3-(4,5-dimethylthiazol-2-yl)-2,5–diphenyltetrazolium bromide (MTT) were from Sigma-Aldrich (St Louis, MO, USA). Lipofectamine™2000 and LysoTracker Blue-DND-99 were supplied by Invitrogen Corporation (Carlsbad, CA, USA). Deoxyribonuclease I (DNase I) was purchased from Gibco BRL (Gaithersburg, MD, USA). CA anhydride, Rhodamine B isothiocyanate (RITC), chlorpromazine, filipin III, and cytochalasin D were from Sigma-Aldrich. pEGFP, transformed in Escherichia coli DH5a, was donated by the First Affiliated Hospital of College of Medicine, Zhejiang University (Hangzhou, People’s Republic of China). All other solvents were of analytical or chromatographic grade.
Cell culture
The HEK-293 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA) and cultured in Dulbecco’s Modified Eagle’s Medium (GibcoBRL, Gaithersberg, MD, USA) with 10% (volume ratio) fetal bovine serum at 37°C under 5% CO2.
Synthesis and characterization of CSO-SA and CA-CSO-SA
CSO-SA was synthesized as previously described with some slight modification.21 CSO-SA was further modified with CA anhydride to obtain CA-CSO-SA via reaction between the amino groups on chitosan and the anhydride on CA anhydride. Briefly, CSO (2.0 g) was dissolved in deionized water (90 mL) and SA (0.6 g) and EDC (1.9 g) were dissolved in ethanol (60 mL). The CSO solution was heated to 60°C under stirring accompanied by dropwise addition of SA solution. The mixtures were stirred at 60°C for 6 hours to obtain CSO-SA. CA-CSO-SA was then obtained by adding the solution containing CSO-SA (10 mg/mL) to the solution containing CA anhydride (2 mg/mL), followed by stirring for 12 hours at 60°C. CSO-SA and CA-CSO-SA were purified by dialysis against water for 3 days (molecular weight cutoff 7,000 kDa), and powders were obtained by lyophilization (FreeZone 2.5 Plus, Labconco Corporation, Kansas City, MO, USA).
CSO-SA and CA-CSO-SA were characterized by 1H-nuclear magnetic resonance (NMR) on an Avance DMX 500 NMR spectrometer (Bruker, Ettlingen, Germany). The CSO-SA and CA-CSO-SA micelles could be easily formed by dispersing CSO-SA and CA-CSO-SA powders into deionized water followed by probe-type ultrasonic treatment. The particle size and zeta potential of the micelles (1.0 mg/mL) were measured by dynamic light scattering (Zetasizer Nano ZS990, Malvern Instruments, Malvern, UK). The morphologies of the nanocomplexes were examined using a JEM-1230 transmission electron microscope (JEOL Ltd, Tokyo, Japan).
The degree of substitution, defined as the number of SA and CA groups per 100 amino groups of CSO, was determined using the trinitrobenzenesulfonic acid method.24 The critical micelle concentration for CSO-SA and CA-CSO-SA was measured by fluorescence using pyrene as a probe. Pyrene (12 mg) was dissolved in acetone (100 mL) and then steadily diluted to a final concentration of 0.0012 mg/mL. Next, 0.5 mL of acetone containing pyrene was added into the test tubes and evaporated naturally at 50°C. A series of sample solutions with CSO-SA and CA-CSO-SA concentrations ranging from 1.0 mg/mL to 5.0 mg/mL was sonicated at room temperature for 30 minutes, and the fluorescence spectra of the solutions were obtained using an F-2500 fluorometer (Hitachi, Tokyo, Japan) at room temperature. The excitation wavelength was 337 nm and the slit openings were 10 nm (excitation) and 2.5 nm (emission). The critical micelle concentration for CSO-SA and CA-CSO-SA was calculated from the intensity ratio of the first peak (I1, 374 nm) to the third peak (I3, 385 nm) in the pyrene emission spectra.
Formation and characterization of nanocomplexes
pEGFP was selected as the model pDNA. CSO-SA/pDNA and CA-CSO-SA/pDNA nanocomplexes at various weight ratios (CSO-SA to pDNA or CA-CSO-SA to pDNA) were prepared by mixing pDNA with CSO-SA and CA-CSO-SA micelles, respectively, at the desired concentrations and then vortexing for 30 seconds. The resulting nanocomplexes were incubated further at 37°C for 30 minutes before use. The morphology of the nanocomplexes was examined by transmission electron microscopy. The association of pDNA with the polymers was monitored using a gel retardation assay on 1% agarose gel stained with ethidium bromide (0.5 mg/mL). Electrophoresis was performed at 100 V for 40 minutes.
DNase I protection
The CSO-SA/pDNA and CA-CSO-SA/pDNA nanocomplexes (containing 1 μg of pDNA) were incubated for 30 minutes at room temperature, after which DNase I (10 U) was added. As a control, naked pDNA (1 μg) was also treated with DNase I under the same conditions. After 30 minutes of incubation, the samples were treated with 4 μL of ethylenediaminetetraacetic acid (250 mM) for 10 minutes to terminate the activation of DNase I, then 5 μL heparin solution (5 wt%) was added and incubated for 30 minutes to replace the pDNA from nanocomplexes. The pDNA released from the nanocomplexes was assessed by agarose gel electrophoresis. Electrophoresis was carried out at 100 V for 40 minutes in Tris-acetate buffer. CSO-SA/pDNA, CA-CSO-SA/pDNA, and naked pDNA without addition of DNase I were also treated under the same conditions and used as a comparison.
Cytotoxicity assay
HEK-293 cells were seeded in 96-well culture plates at a density of 1×104 cells per well in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum. The cytotoxicity of CSO-SA and CA-CSO-SA at various concentrations was evaluated by determining the viability of HEK-293 cell after 48 hours of incubation. Two days later, MTT was added and incubated for 4 hours. Next, the MTT solution was removed and dimethyl sulfoxide was added to the wells. The absorption representing cell viability was detected using a Model 680 microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA) at a wavelength of 570 nm. The cytotoxicity of CSO-SA/pDNA and CA-CSO-SA/pDNA at various weight ratios was also examined using the abovementioned methods.
In vitro transfection
HEK-293 cells were seeded on a 24-well plate at a density of 1×105 cells per well and incubated for 24 hours. The cell culture medium was replaced with Dulbecco’s Modified Eagle’s Medium prior to adding the CSO-SA/pDNA and CA-CSO-SA/pDNA nanocomplexes. The cells were visualized after 72 hours of incubation using a DMI 4000B fluorescence microscope (Leica, Nussloch, Germany). For the quantitative assay, the cells were trypsinized, suspended in 0.1 M ice-cold phosphate-buffered saline (pH 7.4), and investigated using an FC500MCL flow cytometer (Beckman Coulter Inc., Fullerton, CA, USA). The transfection efficiency was calculated as the percentage of green fluorescence-expressing cells per total tested cells. Cells treated with naked pDNA and Lipofectamine 2000/pDNA complexes were used as the negative and positive controls, respectively.
Cellular uptake measurements
The CSO-SA and CA-CSO-SA were labeled with RITC for observation of fluorescence. In brief, the solution containing CSO-SA and CA-CSO-SA (1 mg/mL) was incubated with RITC (2 mg/mL) in the dark for 12 hours, and then dialyzed for purification. RITC-labeled CSO-SA/pDNA and CA-CSO-SA/pDNA were prepared by mixing pDNA and RITC-labeled CSO-SA and CA-CSO-SA, respectively. HEK-293 cells were seeded in 12-well plates at 2×105 cells/well and incubated for 24 hours. Next, the cells were rinsed twice with phosphate-buffered saline and replenished with 2 mL of fresh culture medium containing RITC-labeled CSO-SA/pDNA or CA-CSO-SA/pDNA. After incubation for 4 and 24 hours, the cell nuclei were stained with Hoechst 33342. The cells were then fixed in 4% paraformaldehyde, rinsed twice in phosphate-buffered saline, and examined under an IX81-FV1000 confocal laser scanning microscope (Olympus, Tokyo, Japan). For quantification, the cells were detached using trypsin, resuspended in phosphate-buffered saline, and analyzed by flow cytometry.
Transfection inhibition assay
HEK-293 cells were seeded in 12-well plates at 2×105 cells/well and incubated for 24 hours. The cells were preincubated with a series of inhibitors at concentrations known to have no significant cytotoxic effect. Briefly, the cells were pretreated with Dulbecco’s Modified Eagle’s Medium containing endocytic inhibitors, including chlorpromazine 10 μg/mL (to inhibit formation of clathrin vesicles),25,26 filipin III 5 μg/mL (to inhibit caveolae-mediated endocytosis),27 and cytochalasin D 5 μg/mL (to inhibit macropinocytosis)28 for one hour at 37°C, followed by incubation with CSO-SA/pDNA or CA-CSO-SA/pDNA for 72 hours. The cells were then observed under a fluorescence microscope, and their fluorescence intensities were assessed by flow cytometry. For comparison, cells that had not been treated with these inhibitors were also incubated with CSO-SA/pDNA or CA-CSO-SA/pDNA under the same conditions. Their fluorescence intensities were then determined and expressed as 100%.
Intracellular trafficking
HEK-293 cells were seeded at 5×104 cells/well in 35 mm glass-bottomed dishes. After 24 hours, the cells were incubated with RITC-labeled CSO-SA/pDNA or CA-CSO-SA/pDNA for 12 and 24 hours. The acidic late endosomes and lysosomes were stained using a LysoTracker Blue-99 kit (75 nM) according to the manufacturer’s protocol. The cells were then washed with ice-cold phosphate-buffered saline and observed by confocal laser scanning microscopy.
Statistical analysis
All experiments were repeated in triplicate unless otherwise stated. Student’s t-test was used for statistical analysis to compare the difference between the two groups, with P<0.05 considered to be statistically significant.
Results
Synthesis and characterization of CSO-SA and CA-CSO-SA
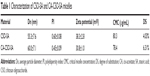
The synthesis of CSO-SA and CA-CSO-SA is shown in Figure 1A and confirmed by H1-NMR in Figure 1B. The peak at about 1.0 ppm is attributed to the methylene and methyl hydrogen of SA in the CSO-SA and CA-CSO-SA compared with CSO. For CA-CSO-SA, the protons at 6.5 ppm are assigned to the protons of the CA group. As shown in Table 1, the substitution degree of amino groups in CA-CSO-SA measured by trinitrobenzenesulfonic acid was 6.31%, while the substitution degree of amino groups in CSO-SA was 4.03%. The critical micelle concentration for CSO-SA and CA-CSO-SA in pure water was 80.3 μg/mL and 78.4 μg/mL, respectively (Table 1), indicating that both materials had excellent self-assembling properties in aqueous medium. The micelles formed by CSO-SA and CA-CSO-SA were positively charged due to the amino group on their surface (38.3±2.8 mV and 30.8±1.0 mV, respectively). Compared with CSO-SA micelles, the zeta potential of the CA-CSO-SA micelles was significantly decreased (P<0.05) because of the carboxyl group in CA. The mean diameter of the CSO-SA and CA-CSO-SA micelles was 55.3±7.6 nm and 60.0±7.1 nm (Table 1), respectively, as determined by dynamic light scattering, and corresponded to the values obtained on transmission electron microscopy (Figure 2A). Clearly, conjugation of CA did not significantly affect the diameter of the CSO-SA micelles. Interestingly, on transmission electron microscopy, the micelles formed by CSO-SA and CA-CSO-SA had the appearance of irregular spherical aggregates.
Formation and characterization of nanocomplexes
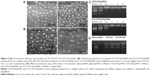
The nanocomplexes were prepared easily by mixing CSO-SA or CA-CSO-SA with pDNA followed by vortexing. It was found that the CA-CSO-SA/pDNA nanocomplexes had a significant larger diameter than the CSO-SA/pDNA nanocomplexes (Figure 2B). A possible explanation for this is that the electrostatic interaction between pDNA and CA-CSO-SA was reduced due to the decreased positive potential of the CA-CSO-SA micelles, and induced loosening of the nanocomplexes. The ability of pDNA and CSO-SA or CA-CSO-SA to bind was assessed by monitoring the electrophoretic mobility of pDNA with agarose gel electrophoresis (Figure 2C). Gradual pDNA retardation was observed as the weight ratio in the nanocomplexes increased. For CA-CSO-SA/pDNA, the migration capability of pDNA was found to be completely hindered at a weight ratio of 2 and upwards, indicating neutralization of the charge on DNA by CA-CSO-SA. For CSO-SA/pDNA, complete DNA retardation was induced beyond a weight ratio of 1. These results indicate that CA-CSO-SA has less binding ability with pDNA than does CSO-SA. The protective effect of CSO-SA and CA-CSO-SA to pDNA against DNase I is shown in Figure 2D. Naked pDNA was completely digested, with no bands observed. In contrast, clear migration bands of pDNA from CSO-SA/pDNA or CA-CSO-SA/pDNA at a weight ratio of 2.5 were noted via substitution of heparin. These results suggest that both CSO-SA and CA-CSO-SA can protect pDNA effectively, which would be very important for safe delivery of therapeutic DNA to its target.
Cytotoxicity assay
The cytotoxicity of CSO-SA and CA-CSO-SA was evaluated in HEK-293 cells using the MTT assay. As shown in Figure 3A, both CSO-SA and CA-CSO-SA had minimal cytotoxicity at a low concentration (10 μg/mL) but their cytotoxicity increased with increasing concentrations. The IC50 for CSO-SA in HEK-293 cells was 0.3 mg/mL. However, for CA-CSO-SA, more than 50% of cells survived, even at the highest concentration used (1.0 mg/mL), suggesting that CA-CSO-SA could have better biosafety than CSO-SA. As shown in Figure 3B, CSO-SA/pDNA or CA-CSO-SA/pDNA at weight ratios of 2.5–15 induced almost no cell death (>90% cell viability), while nanocomplexes of Lipofectamine 2000/pDNA (2 μg Lipofectamine per 1 μg pDNA) showed slight cytotoxicity at the same pDNA concentration (~75% cell viability). These results suggest that CSO-SA/pDNA is more biocompatible than Lipofectamine/pDNA.
Gene transfection efficiency
The gene transfection ability of CSO-SA/pDNA and CA-CSO-SA/pDNA was investigated in HEK-293 cells in the presence of 10% fetal bovine serum. As shown in Figure 4A, CA-CSO-SA/pDNA showed more green fluorescent expression than CSO-SA/pDNA at the same weight ratio. In order to directly compare gene expression for CSO-SA/pDNA and CA-CSO-SA/pDNA, the cells were analyzed by fluorescence-activated cell sorting. The transfection efficiency of CA-CSO-SA/pDNA at weight ratios (CA-CSO-SA to pDNA) of 2.5, 5, and 10 was 18%, 37%, and 24%, respectively, whereas the transfection efficiency of CSO-SA/pDNA was below 18% for all tested weight ratios (Figure 4B). These results indicate that CA-CSO-SA could be a significantly more efficient gene carrier than CSO-SA. CA-CSO-SA/pDNA showed the highest transfection efficiency at a weight ratio of 5, which is a little higher than that of Lipofectamine/pDNA (33%) under optimized conditions according to the manufacturer’s protocol. There was almost no green fluorescent expression in cells treated with naked pDNA.
Internalization and endocytosis pathway
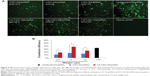
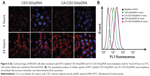
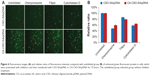
The fluorescent signal, indicating the amount of cellular uptake, increased with longer incubation times for both CA-CSO-SA/pDNA and CSO-SA/pDNA (Figure 5A). CA-CSO-SA/pDNA was taken up more slowly by cells than CSO-SA/pDNA at shorter incubation times (eg, 4 hours), but there was no significant difference in cell uptake between CA-CSO-SA/pDNA and CSO-SA/pDNA at longer incubation times (eg, 24 hours) according to the flow cytometry assay (Figure 5B). The decreased CA-CSO-SA/pDNA uptake rate with short incubation times might be attributable to the lower zeta potential and increased hydrophobicity of CA-CSO-SA, which inhibits electrostatic interaction between the nanocomplexes and the cell membrane.29 The mechanism via which the nanocomplexes were taken up was investigated further by treating cells with chemical inhibitors of clathrin-mediated endocytosis, caveolae-mediated endocytosis, and macropinocytosis. Inhibition of clathrin-mediated endocytosis was investigated using chlorpromazine, which causes clathrin to accumulate in late endosomes, thereby inhibiting coated pit-mediated endocytosis. Filipin inhibits caveolae-mediated endocytosis,30 and cytochalasin D is known to inhibit actin polymerization and membrane ruffling, which is involved in macropinocytosis.31 Addition of each inhibitor decreased the transfection efficiency of both CSO-SA/pDNA and CA-CSO-SA/pDNA (Figure 6A). The gene transfection efficiency in cells treated with chlorpromazine was reduced more than in cells treated with the other inhibitors (up to 50% and 40% for CSO-SA/pDNA and CA-CSO-SA/pDNA, respectively, Figure 6B). Our inhibition studies suggest that internalization of CA-CSO-SA/pDNA and CSO-SA/pDNA is mediated by three cellular uptake mechanisms, ie, clathrin-mediated endocytosis, caveolae-mediated endocytosis, and macropinocytosis, with the clathrin-mediated pathway playing the most important role.
Intracellular trafficking
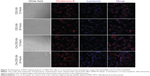
It is known that successful escape from the endosome is crucial for gene carriers to be able to improve gene transfection efficiency. To compare the ability of CSO-SA/pDNA and CA-CSO-SA/pDNA to escape the endosome, the intracellular distribution of each nanocomplex was investigated by confocal laser scanning microscopy (Figure 7). Late endosomes/lysosomes were stained with Lyso-Tracker Blue. After 12 hours of incubation, most of the CSO-SA/pDNA and CA-CSO-SA/pDNA was colocalized within late endosomes/lysosomes. After 24 hours of incubation, the location of most of the red signal for CA-CSO-SA/pDNA did not match that of the blue signal, indicating that most of the CA-CSO-SA/pDNA had escaped from the late endosomes/lysosome compartment. However, it was found that most of the CSO-SA/pDNA was still entrapped in late endosomes/lysosome compartment after the same incubation time. The ability of CA-CSO-SA/pDNA to escape more rapidly from late endosomes/lysosomes is likely attributable to the increased number of carboxyl groups in CA-CSO-SA.
Discussion
Over the past decade, chitosan and its derivatives have emerged as promising vehicles for nonviral gene delivery. However, the transfection efficiency shown by chitosan-based gene carriers is insufficient for practical application, so researchers have attempted to improve the transfection efficiency of chitosan and its derivatives.7,8 CA-CSO-SA, synthesized to have the advantages of protecting DNA, excellent internalization, and rapid escape from the endosome, have been shown to be a promising gene delivery system with high transfection efficiency.
The formation of stable nanocomplexes of CA-CSO-SA and pDNA under physiological conditions is a prerequisite for efficient gene delivery. It was clear that DNA could easily be compacted into nanocomplexes with CSO-SA or CA-CSO-SA micelles. The capacity of CA-CSO-SA to complex pDNA was slightly decreased by introduction of carboxyl moieties. However, CA-CSO-SA could still protect pDNA from degradation by nucleases known to be abundant in serum and the extracellular matrix if the weight ratio of CA-CSO-SA to pDNA was over 2.5 (Figure 2D). It is also known that the physical stability of gene delivery systems in serum is a very important factor affecting their use in vivo.32 High transfection efficiency was obtained under serum conditions (Figure 4), suggesting that CA-CSO-SA may be useful for transfection in vivo.
High cellular uptake was also an important factor for gene transfection. Excellent uptake of CSO-SA/pDNA by tumor cells has been reported, and this is attributed to the specific structure of the micelles formed by CSO-SA.21 CSO-SA micelles have multiple “minor cores” on their surface, which can insert into the membrane of the cell, causing hydrophobic interactions with components of the cell membrane, resulting in perturbation and fusion of the membrane, thereby facilitating endocytosis of the micelles.33 In our study, cellular uptake of CA-CSO-SA/pDNA was again excellent, with uptake that was similar to that of CSO-SA/pDNA after 24 hours of incubation (Figure 5), suggesting that CA-CSO-SA/pDNA and CSO-SA/pDNA could share the same mechanism of cell internalization. Given that the intracellular fate of gene carriers is determined by their route of uptake, studies of the mechanisms of cell internalization has been gaining in importance.34 So far, the endocytosis pathways for CSO-SA/pDNA have not been clear. In this work, we investigated the contribution of three internalization pathways for CSO-SA/pDNA and CA-CSO-SA/pDNA, ie, clathrin-mediated endocytosis, caveolae-mediated endocytosis, and macropinocytosis. Our data indicate that clathrin-mediated endocytosis, which provides entry into endolysosomes, plays the most important role in cellular internalization of both these nanocomplexes. Further, addition of carboxyl groups did not alter the internalization pathway for CA-CSO-SA/pDNA.
Intracellular trafficking of the nanocomplexes after uptake into the cell has a strong influence on transfection efficiency. Chitosan-based nanocomplexes transit through lysosomes for a relatively long period of time prior to transfection.35 Ideal vectors should be able to escape easily from endolysosomes to avoid degradation of pDNA by acidic components and enzymes.36 We found that CA-CSO-SA/pDNA could escape from endolysosomes and be released into the cytoplasm more rapidly than CSO-SA/pDNA. The addition of a carboxyl group onto CA-CSO-SA may be a key factor. Water-soluble polymers, which contain many carboxylic acid groups, have been shown to have a membrane-destabilizing effect under weakly acidic conditions, and can cause disruption of the endosomal membrane and facilitate transport of the nanocomplexes from the endosome to the cytosol.37,38 Anionic polymers, such as polyacrylic acid and amphiphilic acidic peptides, have been investigated for their ability to enhance transfection efficiency.14,39,40 Thus, introduction of CA moieties onto CSO-SA would be expected to have an enhancing effect on gene transfection efficiency. A possible mechanism is the carboxyl in CA-CSO-SA being in a state of dissociation under physiological conditions and protons being able to combine under low pH conditions in endolysosomes. The change in the ionization state of the carboxyl group thus leads to a conformational alteration within the nanocomplexes, promoting their escape from endolysosomes.
The toxicity of nonviral gene carriers also needs to be addressed. In this study, both types of nanocomplex had little cytotoxicity in HEK-293 cells under transfection conditions. The cytotoxicity of CA-CSO-SA was significantly decreased compared with that of CSO-SA. The reason for this may be that the carboxyl group in CA-CSO-SA partially shields the positive charge of the material. Further, it have been widely reported that carboxylate groups are less toxic than amine groups.41 These results are in accordance with those previously reported for gene carriers.42 With our growing and more comprehensive understanding of polymer-based gene delivery, it is likely that these gene delivery systems will become an important tool in human gene therapy.
Conclusion
In this study, CA CSO-SA was investigated as a nonviral vector for gene delivery. Our data demonstrate that CA-CSO-SA could efficiently protect pDNA from degradation and be successfully taken up into cells by clathrin-mediated endocytosis, caveolae-mediated endocytosis, and macropinocytosis. Our data also indicate that CA-CSO-SA/pDNA escaped from the endosome efficiently and that the rate of escape was significantly more rapid than that of CSO-SA/pDNA. The transfection efficiency of CA-CSO-SA/pDNA in HEK-293 cells at the optimal weight ratio was 37%, showing a one-fold increase compared with CSO-SA/pDNA. Taken together, the excellent transfection efficiency of CA-CSO-SA/pEGFP can be attributed to protection of DNA, excellent cellular uptake, and efficient escape from the endosome. Our results suggest that CA-CSO-SA is an efficient, safe, and promising material for gene delivery.
Acknowledgment
The authors are grateful for the financial support of the National Nature Science Foundation of China (81273442) and the Zhejiang Provincial Program for the Cultivation of High-Level Innovative Health Talents.
Disclosure
The authors report no conflicts of interest in this work.
References
Raviña M, Cubillo E, Olmeda D, et al. Hyaluronic acid/chitosan-g-poly (ethylene glycol) nanoparticles for gene therapy: an application for pDNA and siRNA delivery. Pharm Res. 2010;27:2544–2555. | ||
Jones CH, Chen CK, Ravikrishnan A, Rane S, Pfeifer BA. Overcoming nonviral gene delivery barriers: perspective and future. Mol Pharm. 2013;10:4082–4098. | ||
Miyata K, Nishiyama N, Kataoka K. Rational design of smart supramolecular assemblies for gene delivery: chemical challenges in the creation of artificial viruses. Chem Soc Rev. 2012;41:2562–2574. | ||
Mizrahy S, Peer D. Polysaccharides as building blocks for nanotherapeutics. Chem Soc Rev. 2012;41:2623–2640. | ||
Liu Z, Jiao Y, Wang Y, Zhou C, Zhang Z. Polysaccharides-based nanoparticles as drug delivery systems. Adv Drug Deliv Rev. 2008;60:1650–1662. | ||
Saranya N, Moorthi A, Saravanan S, Devi MP, Selvamurugan N. Chitosan and its derivatives for gene delivery. Int J Biol Macromol. 2011;48:234–238. | ||
Strand SP, Lelu S, Reitan NK, de Lange Davies C, Artursson P, Vårum KM. Molecular design of chitosan gene delivery systems with an optimized balance between polyplex stability and polyplex unpacking. Biomaterials. 2010;31:975–987. | ||
Sun B, Zhao R, Kong F, et al. Phosphorylatable short peptide conjugation for facilitating transfection efficacy of CS/DNA complex. Int J Pharm. 2010;397:206–210. | ||
Wang B, He C, Tang C, Yin C. Effects of hydrophobic and hydrophilic modifications on gene delivery of amphiphilic chitosan based nanocarriers. Biomaterials. 2011;32:4630–4638. | ||
Zheng H, Tang C, Yin C. The effect of crosslinking agents on the transfection efficiency, cellular and intracellular processing of DNA/polymer nanocomplexes. Biomaterials. 2013;34:3479–3488. | ||
Mao S, Sun W, Kissel T. Chitosan-based formulations for delivery of DNA and siRNA. Adv Drug Deliv Rev. 2010;62:12–27. | ||
Lai WF, Lin MC. Nucleic acid delivery with chitosan and its derivatives. J Control Release. 2009;134:158–168. | ||
Lundberg P, El-Andaloussi S, Sütlü T, Johansson H, Langel U. Delivery of short interfering RNA using endosomolytic cell-penetrating peptides. FASEB J. 2007;21:2664–2671. | ||
Kyriakides TR, Cheung CY, Murthy N, Bornstein P, Stayton PS, Hoffman AS. pH-sensitive polymers that enhance intracellular drug delivery in vivo. J Control Release. 2002;78:295–303. | ||
Ito T, Yoshihara C, Hamada K, Koyama Y. DNA/polyethyleneimine/hyaluronic acid small complex particles and tumor suppression in mice. Biomaterials. 2010;31:2912–2918. | ||
Liao ZX, Ho YC, Chen HL, Peng SF, Hsiao CW, Sung HW. Enhancement of efficiencies of the cellular uptake and gene silencing of chitosan/siRNA complexes via the inclusion of a negatively charged poly(gamma-glutamic acid). Biomaterials. 2010;31:8780–8788. | ||
Wang C, Luo X, Zhao Y, et al. Influence of the polyanion on the physico-chemical properties and biological activities of polyanion/DNA/polycation ternary polyplexes. Acta Biomater. 2012;8:3014–3026. | ||
Hu FQ, Liu LN, Du YZ, Yuan H. Synthesis and antitumor activity of doxorubicin conjugated stearic acid-g-chitosan oligosaccharide polymeric micelles. Biomaterials. 2009;30:6955–6963. | ||
Hu FQ, Wu XL, Du YZ, You J, Yuan H. Cellular uptake and cytotoxicity of shell crosslinked stearic acid-grafted chitosan oligosaccharide micelles encapsulating doxorubicin. Eur J Pharm Biopharm. 2008;69:117–125. | ||
Du YZ, Lu P, Yuan H, Zhou JP, Hu FQ. Quaternary complexes composed of plasmid DNA/protamine/fish sperm DNA/stearic acid grafted chitosan oligosaccharide micelles for gene delivery. Int J Biol Macromol. 2011;48:153–159. | ||
Hu FQ, Zhao MD, Yuan H, You J, Du YZ, Zeng S. A novel chitosan oligosaccharide-stearic acid micelles for gene delivery: properties and in vitro transfection studies. Int J Pharm. 2006;315:158–166. | ||
You J, Hu FQ, Du YZ, Yuan H. Polymeric micelles with glycolipid-like structure and multiple hydrophobic domains for mediating molecular target delivery of paclitaxel. Biomacromolecules. 2007;8:2450–2456. | ||
Hu F, Chen W, Zhao M, Yuan H, Du YZ. Effective antitumor gene therapy delivered by polyethylenimine-conjugated stearic acid-g-chitosan oligosaccharide micelles. Gene Ther. 2012;20:597–606. | ||
Yan J, Du YZ, Chen FY, et al. Effect of proteins with different isoelectric points on the gene transfection efficiency mediated by stearic acid grafted chitosan oligosaccharide micelles. Mol Pharm. 2013;10:2568–2577. | ||
Douglas KL, Piccirillo CA, Tabrizian M. Cell line-dependent internalization pathways and intracellular trafficking determine transfection efficiency of nanoparticle vectors. Eur J Pharm Biopharm. 2008;68:676–687. | ||
Chiu YL, Ho YC, Chen YM, et al. The characteristics, cellular uptake and intracellular trafficking of nanoparticles made of hydrophobically-modified chitosan. J Control Release. 2010;146:152–159. | ||
Manunta M, Tan PH, Sagoo P, Kashefi K, George AJ. Gene delivery by dendrimers operates via a cholesterol dependent pathway. Nucleic Acids Res. 2004;32:2730–2739. | ||
Gratton SE, Ropp PA, Pohlhaus PD, et al. The effect of particle design on cellular internalization pathways. Proc Natl Acad Sci U S A. 2008;105:11613–11618. | ||
Nel AE, Mädler L, Velegol D, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8:543–557. | ||
Orlandi PA, Fishman PH. Filipin-dependent inhibition of cholera toxin: evidence for toxin internalization and activation through caveolae-like domains. J Cell Biol. 1998;141:905–915. | ||
Perumal OP, Inapagolla R, Kannan S, Kannan RM. The effect of surface functionality on cellular trafficking of dendrimers. Biomaterials. 2008;29:3469–3476. | ||
Wen Y, Guo Z, Du Z, et al. Serum tolerance and endosomal escape capacity of histidine-modified pDNA-loaded complexes based on polyamidoamine dendrimer derivatives. Biomaterials. 2012;33:8111–8121. | ||
You J, Hu FQ, Du YZ, Yuan H, Ye BF. High cytotoxicity and resistant-cell reversal of novel paclitaxel loaded micelles by enhancing the molecular-target delivery of the drug. Nanotechnology. 2007;18:495101. | ||
Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol Rev. 2006;58:32–45. | ||
Thibault M, Nimesh S, Lavertu M, Buschmann MD. Intracellular trafficking and decondensation kinetics of chitosan-pDNA polyplexes. Mol Ther. 2010;18:1787–1795. | ||
Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. | ||
Koyama Y, Ito T, Matsumoto H, et al. Novel poly(ethylene glycol) derivatives with carboxylic acid pendant groups: synthesis and their protection and enhancing effect on non-viral gene transfection systems. J Biomater Sci Polym Ed. 2003;14:515–531. | ||
Stayton PS, Hoffman AS, Murthy N, et al. Molecular engineering of proteins and polymers for targeting and intracellular delivery of therapeutics. J Control Release. 2000;65:203–220. | ||
Thomas JL, Tirrell DA. Polymer-induced leakage of cations from dioleoyl phosphatidylcholine and phosphatidylglycerol liposomes. J Control Release. 2000;67:203–209. | ||
Mahat R, Monera O, Smith L, Rolland A. Peptide-based gene delivery. Curr Opin Mol Ther. 1999;1:226–243. | ||
Yezhelyev MV, Qi L, O’Regan RM, Nie S, Gao X. Proton-sponge coated quantum dots for siRNA delivery and intracellular imaging. J Am Chem Soc. 2008;130:9006–9012. | ||
Jevprasesphant R, Penny J, Attwood D, McKeown NB, D’Emanuele A. Engineering of dendrimer surfaces to enhance transepithelial transport and reduce cytotoxicity. Pharm Res. 2003;20:1543–1550. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.