Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Coupling of a bifunctional peptide R13 to OTMCS-PEI copolymer as a gene vector increases transfection efficiency and tumor targeting
Authors Lv H, Zhu Q, Liu K, Zhu M, Zhao W, Mao Y, Liu K
Received 25 December 2013
Accepted for publication 21 January 2014
Published 11 March 2014 Volume 2014:9(1) Pages 1311—1322
DOI https://doi.org/10.2147/IJN.S59726
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Hui Lv,1,* Qing Zhu,1,* Kewu Liu,2 Manman Zhu,1 Wenfang Zhao,1 Yuan Mao,1 Kehai Liu1
1Department of Biopharmaceutics, College of Food Science and Technology, Shanghai Ocean University, Shanghai, People's Republic of China; 2Heilongjiang Forest By-Product and Speciality Institute, Mudanjiang, People's Republic of China
*These authors contributed equally to this work
Background: A degradable polyethylenimine (PEI) derivative coupled to a bifunctional peptide R13 was developed to solve the transfection efficiency versus cytotoxicity and tumor-targeting problems of PEI when used as a gene vector.
Methods: We crossed-linked low molecular weight PEI with N-octyl-N-quaternary chitosan (OTMCS) to synthesize a degradable PEI derivative (OTMCS-PEI), and then used a bifunctional peptide, RGDC-Tat (49–57) called R13 to modify OTMCS-PEI so as to prepare a new gene vector, OTMCS-PEI-R13. This new gene vector was characterized by various physicochemical methods. Its cytotoxicity and gene transfection efficiency were also determined both in vitro and in vivo.
Results: The vector showed controlled degradation and excellent buffering capacity. The particle size of the OTMCS-PEI-R13/DNA complexes was around 150–250 nm and the zeta potential ranged from 10 mV to 30 mV. The polymer could protect plasmid DNA from being digested by DNase I at a concentration of 23.5 U DNase I/µg DNA. Further, the polymer was resistant to dissociation induced by 50% fetal bovine serum and 400 µg/mL sodium heparin. Compared with PEI 25 kDa, the OTMCS-PEI-R13/DNA complexes showed higher transfection efficiency both in vitro and in vivo. Further, compared with OTMCS-PEI, distribution of OTMCS-PEI-R13 at tumor sites was markedly enhanced, indicating the tumor-targeting specificity of R13.
Conclusion: OTMCS-PEI-R13 could be a potential candidate as a safe and efficient gene delivery carrier for gene therapy.
Keywords: nonviral gene vector, polyethylenimine, R13, transfection efficiency, tumor-targeting
Introduction
Cancer is becoming a scourge in modern society. Traditional treatments such as surgery, radiotherapy, and chemotherapy are not always effective at curing cancers, so there is an urgent need for new strategies.1 Based on genetic engineering, gene therapy is becoming a new method of curing cancer. The basis of gene therapy is insertion of therapeutic genes into target cells via gene vectors2 and key to this is finding an ideal gene delivery vector. An ideal gene vector needs to be able to condense DNA effectively, be stable in body fluids, target specific cells, cross membranes, and release its cargo efficiently.3
At present, two kinds of gene vectors are available, ie, viral vectors and nonviral vectors. Viral vectors are seriously limited by a number of disadvantages, including immunogenicity, poor loading capacity, and oncogenic effects.4 In contrast, nonviral gene vectors are noninfectious, nonimmunogenic, and can carry large amounts of genetic material.5–7 As a result of these characteristics, nonviral gene vectors have been paid more and more attention.8
Among the nonviral vectors, cationic polymers have been widely studied in gene delivery research. Recently, delivery of nucleic acids into cells by cationic polymers has attracted interest in the field of nonviral gene therapy, owing to their easy production, safety, structural diversity, and nonimmunogenicity.9 One of the most effective and widely studied synthetic nonviral gene delivery vectors is the polyethylenimine (PEI) polycation, which has recently been used for the design of DNA delivery vehicles.
PEI and its derivatives are being extensively investigated because of their “proton sponge” effect.10,11 PEI has an abundance of positive charges that can condense DNA via electrostatic interaction into compact complexes that can break through various barriers to the nuclei of target cells.12 Because DNA is condensed, it is not easily degraded by nucleases or gathered as a precipitate, which results in high transfection efficiency.13,14 However, it has been reported that PEI/DNA complexes have three unresolved problems. First, the molecular weight of PEI has a strong influence on the transfection efficiency and cytotoxicity. PEI with a high molecular weight has high transfection efficiency but also high cytotoxicity. On the other hand, PEI with a low molecular weight has low cytotoxicity, but low transfection efficiency.11 Secondly, there is an inconsistency between PEI’s stability in body fluid and cellular uptake.15 Third, delivery of genes by PEI lacks satisfactory specificity for tumor cells because there is no binding selectivity between the positively charged polycations and the negatively charged cells in the body.15
It has been shown that PEI derivatives obtained by cross-linking low molecular weight PEI with degradable materials have higher transfection efficiency and lower cytotoxicity.16 Chitosan, obtained by alkaline N-deacetylation of chitin, is one of the most widely used natural cationic polysaccharides.17 It shows particularly high biocompatibility and fairly low cytotoxicity,18 and therefore can be used as a gene vector.19,20 Nevertheless, chitosan is insoluble at physiological pH and lacks charge. These drawbacks limit its use as a gene vector.21
In light of the above considerations, we synthesized N-octyl-N-quaternary chitosan (OTMCS) and cross-linked this amphiphilic chitosan with low molecular weight PEI, thereby obtaining a novel degradable OTMCS-PEI derivative with high transfection efficiency and low cytotoxicity. Our previous work has demonstrated that OTMCS-PEI can greatly reduce the cytotoxicity of PEI while ensuring higher transfection efficiency, thus solving the problem of efficiency versus cytotoxicity.22
Receptor-mediated tumor targeting is another increasingly important area in nonviral gene delivery research. Integrins are important cell surface receptors. They are composed of α (120–185 kDa) and β (90–110 kDa) subunits and can recognize a number of proteins as receptors. To date, over 20 integrins have been identified. Among them is the αvβ3 receptor, which is highly expressed on tumor cells and angiogenic tumor blood vessels but is rarely expressed on normal cells. Thus, αvβ3 could be a promising target for antitumor therapy.23
The arginine-glycine-aspartate-cysteine (RGDC) peptide is a key binding moiety that binds specifically to αvβ3 integrin receptors.24 Cell-penetrating peptides contain 7–30 amino acids, which can transport various molecules across the plasma membrane rapidly and target the cell nucleus. The transduction domain, ie, the region conveying the cell-penetrating peptides, appears to be confined to a small stretch of basic amino acids with the sequence RKKRRQRRR and is known as TAT (49–57).25 Therefore, we used an RGDC peptide in conjunction with TAT to yield a new chimeric peptide, RGDC-TAT, known as R13. Our previous work has demonstrated that R13 can target αvβ3 receptors, which are expressed abundantly on tumor cells, and carry cargo molecules into such cells.26 Therefore, we used R13 to modify OTMCS-PEI in order to improve its tumor-targeting specificity and increase cellular uptake of genes.
In this experiment, we first linked low molecular weight PEI with OTMCS to obtain degradable OTMCS-PEI, then used the tumor-targeting peptide RGDC in conjunction with TAT (49–57) to yield a bifunctional peptide, R13, and finally coupled R13 to OTMCS-PEI so as to prepare a new nonviral gene delivery vector, ie, OTMCS-PEI-R13. This new gene vector was characterized in terms of its chemical structure and biophysical parameters. Its cytotoxicity and gene transfection efficiency were determined both in vitro and in vivo. The purpose of this research was to reduce the cytotoxicity of PEI, improve its transfection efficiency and tumor-targeting ability, increase the cellular uptake of genes, and enhance the efficacy of gene therapy in cancer.
Materials and methods
Materials
Branched PEI (molecular weight 2,000), heparin, N-succinimidyl-4-(N-maleimido-methyl) cyclo-hexane-carboxylate (SMCC), and 3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) were obtained from Sigma-Aldrich (St Louis, MO, USA). Chitosan, benzene, dichloromethane, triphosgene, N-hydroxysuccinimide, triethylamine, ethyl acetate, and absolute ethyl alcohol were purchased from Sinopharm Chemical Reagent Co, Ltd (Shanghai, People’s Republic of China). A dialysis bag (molecular weight cutoff 7,000) was purchased from Shanghai Gene Star Co, Ltd (Shanghai, People’s Republic of China). DNase I was purchased from the Worthington Company (Lakewood, NJ, USA). Roswell Park Memorial Institute 1640 culture medium and fetal bovine serum were purchased from Invitrogen (Carlsbad, CA, USA). A luciferase assay system for the in vitro transfection assay and a pGL3-control vector with SV-40 promoter and enhancer driving firefly (Photinus pyralis) luciferase were obtained from Promega (Madison, WI, USA). The plasmid encoding enhanced green fluorescent protein (pEGFP-N2) was kindly provided by the Institute of Life Science and Technology at Tongji University (Shanghai, People’s Republic of China). The plasmids were amplified using Escherichia coli DH5α and prepared using the Qiagen End-free Plasmid Mega Kit (Qiagen GmbH, Hilden, Germany). The purity of the purified and concentrated DNA was determined by measuring its ultraviolet absorbance at 260 nm and 280 nm. The Arg-Gly-Asp-Cys-Arg-Lys-Arg-Gln-Arg-Arg-Arg oligopeptide (R13) was synthesized by GL Biochem (Shanghai, People’s Republic of China), and the amino acid sequence was confirmed by mass spectroscopy.
Synthesis of OTMCS-PEI-R13
OTMCS and OTMCS-PEI were synthesized according to the method described in our previous work.22 R13 was conjugated with OTMCS-PEI using SMCC as a heterobifunctional agent. Conjugation was performed as described in the manufacturer’s instructions. SMCC was dissolved in dimethylsulfoxide at a concentration of 3.33 mg/mL and OTMCS-PEI was dissolved in 0.1 M phosphate-buffered saline at a concentration of 10 mg/mL, respectively. Next, the SMCC solution was slowly added to the OTMCS-PEI solution at molar ratios of 10:1 and 2:1. The reaction mixture was incubated for 30 minutes at room temperature with gentle shaking. The excess nonreacted cross-linker was removed by gel chromatography (Pharmacia, Piscataway, NJ, USA). R13 was then dissolved in 0.1 M phosphate-buffered saline at a concentration of 10 mg/mL and reacted with maleimide OTMCS-PEI solution at molar ratios of 10:1 and 2:1. The reaction mixture was stirred at 4°C overnight in the dark and then lyophilized after removal of the nonreacted R13 by ultrafiltration with an Amicon Ultra-4 centrifugal filter (EMD Millipore Corporation, Billerica, MA, USA). The products were named OTMCS-PEI-R13-h and OTMCS-PEI-R13-l, respectively.
Characterization of OTMCS-PEI-R13
First, 10 mg of OTMCS-PEI-R13 was dissolved in 0.6 mL of deuterium oxide (D2O) in a nuclear magnetic resonance (NMR) tube, and the 1H NMR spectrum was recorded using a 300 mHz spectrometer (Varian Medical Systems, Palo Alto, CA, USA) at room temperature.
The molecular weight and distribution of the polymer was measured by gel permeation chromatography with multiangle laser light scattering (GPC-MALLS; LC-20AD; Shimadzu Corp, Kyoto, Japan) and a laser wavelength of 690 nm, using a TSK-GEL G5000PWXL column (temperature 40°C) operated at a flow rate of 0.4 mL per minute. Ammonium acetate 0.2 M was used as the mobile phase.
Degradation of OTMCS-PEI-R13
Degradation of OTMCS-PEI-R13 was estimated by measuring its molecular weight. First, 0.5 g of the polymer was dissolved in 10 mL of phosphate-buffered saline (0.1 M, pH 7.4) and then incubated at 37°C with shaking at 100 rpm. After incubation for different periods of time, the polymer solutions were lyophilized and the molecular weights of the lyophilized samples were measured by GPC-MALLS at a 690 nm laser wavelength.
Buffering capacity of OTMCS-PEI-R13
OTMCS-PEI-R13 polymer solutions were prepared in a 50 mL flask (0.2 mg/mL, 30 mL). Pure water was used as a control. After adjusting the initial pH to 10.0 using 0.1 M NaOH, 25 μL increments of 0.1 M HCl were added dropwise into the solution and the pH was measured by a pH electrode at the same time. Variation in pH was recorded from 10.0 to 3.0.
Preparation of OTMCS-PEI-R13/ DNA complexes
The charge ratio was defined as the ratio of amino groups to phosphate groups. The charge ratios of OTMCS-PEI-R13/DNA were controlled by regulating the weight ratios of OTMCS-PEI-R13 and DNA. A polymer solution and a DNA solution were mixed to form self-assembling complexes with the desired weight/weight (w/w) ratios. The complexes were allowed to stand at 4°C for 30 minutes before use in subsequent experiments.
Measurement of particle size and zeta potential, and morphologic observation
The particle size and zeta potential of the polymer/DNA complexes were measured with an electrophoretic light-scattering spectrophotometer (Zetasizer Nano ZS90, MAN0317 Issue 5.0; Malvern Instruments, Malvern, UK) with 90° scattering angles. The complexes were prepared at designed w/w ratios and incubated at 4°C for 30 minutes before size and zeta potential measurements. All the experiments were conducted in triplicate.
The OTMCS-PEI-R13/DNA complexes were prepared as described earlier. The DNA concentration was 50 μg/mL and the w/w ratio of the polymer to DNA was 10. A drop of the complex solution was put on a copper grid. After the sample was dried, the micelle morphology was observed by transmission electron microscopy (JEM 2100F; JEOL Ltd, Tokyo, Japan).
Agarose gel retardation assay
Before electrophoresis, 2 μL of 6× loading buffer was added to 10 μL of the complex solution (250 ng of plasmid DNA [pDNA]). Electrophoresis was carried out on 1% agarose gel at 120 V for about 40 minutes in Tris-acetate-ethylenediamine tetraacetic acid (EDTA) buffer. The gel was then stained with ethidium bromide for 15 minutes and photographed using an ultraviolet illuminator to visualize the DNA migration patterns.
Resistance to DNase I digestion
Designated amounts of DNase I solution were added into 10 μL of the complex solution (containing 250 ng of pDNA) in 0.5 mL Eppendorf tubes and incubated at 37°C for 15 minutes. Next, 2 μL of 250 mM EDTA was added to each tube and incubated at room temperature for 10 minutes in order to inactivate DNase I. Next, 10 μL of 10 mg/mL sodium heparin solution was added and incubated at room temperature for 2 hours to dissociate the complex completely. Electrophoresis was then performed to evaluate the ability of OTMCS-PEI-R13/DNA to resist digestion by DNase I.
Resistance to serum and heparin
First, 10 μL of the prepared complex solution (250 ng of pDNA) was put into 0.5 mL Eppendorf tubes. Different volumes of fetal bovine serum solution or 2 μL of different concentrations of sodium heparin solution were added and incubated at 37°C for 30 minutes. Electrophoresis was then performed to evaluate the ability of OTMCS-PEI-R13/DNA to resist fetal bovine serum and sodium heparin.
Cytotoxicity assay
The cytotoxic effect of OTMCS-PEI-R13 was assessed in HeLa cells using the MTT assay. HeLa cells were seeded at a density of 5,000 cells per well in 200 μL of growth medium in 96-well plates and incubated to 80% confluence. The culture medium was then replaced with 200 μL of fresh serum-free medium containing various concentrations of OTMCS-PEI-R13, PEI 25 kDa, or PEI 2 kDa and incubated for a further 72 hours. Next, the medium was replaced by 20 μL of MTT (5 mg/mL) and 180 μL of fresh growth medium, and incubated for 4 hours. The MTT solution was then replaced by 150 μL of dimethyl sulfoxide and agitated for 10 minutes. Absorbance at 570 nm was determined using an enzyme-linked immunosorbent assay plate reader (Model 680; Bio-Rad, Hercules, CA, USA). Cell viability (%) was calculated using these data and the following equation:26
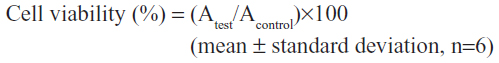
where Atest is the absorbance value for OTMCS-PEI-R13-treated or PEI-treated cells and Acontrol is the absorbance value for untreated cells.
Gene transfection efficiency in vitro
The transfection efficiency of OTMCS-PEI-R13 was evaluated in HeLa cells using pEGFP-N2 and pGL3-control. The cells were seeded in 500 μL of growth medium per well in 24-well plates at a density of 1×105 and incubated to 80% confluence. Before gene transfection, the medium was replaced with 100 μL of complex solution at various weight ratios and 400 μL of serum-free Roswell Park Memorial Institute 1640 medium. The cells were incubated for 4 hours at 37°C in a 5% CO2 atmosphere. The medium was then replaced with 500 μL of medium containing 10% fetal bovine serum and incubated for 48 hours. Next, an inverted fluorescent AE-31 microscope (Motic Corporation, Wetzlar, Germany) was used to observe pEGFP-N2 expression. Transfection efficiency was then determined by flow cytometry (BD FaCSAria™; BD Biosciences, Franklin Lakes, NJ, USA) after the cells were trypsinized, centrifuged, and resuspended in phosphate-buffered saline. The data were analyzed using CellQuest software (Becton Dickinson, Franklin Lakes, NJ, USA).
The luciferase assay was conducted according to the manufacturer’s specifications. The medium was replaced with 100 μL of cell culture lysis reagent and shaken for 30 minutes. After mixing with the substrate, the activity of luciferase was measured using a TD-20/20 luminometer (Promega). A PP1001 bicinchoninic acid protein assay kit (Bioteke Corporation, Beijing, People’s Republic of China) was used to measure protein content. The transfection efficiency for the pGL3-control was calculated as relative light units against the corresponding protein content.
Gene transfection efficiency in vivo
The in vivo gene transfection study was conducted using male BALB/C mice (age 4–5 weeks, weight 18–22 g) purchased from National Rodent Laboratory Animal Resources, Shanghai Branch (Shanghai, People’s Republic of China). The mice were maintained under specific pathogen-free conditions. All animal procedures were approved by the Committee for Animal Research of Shanghai Ocean University, People’s Republic of China, and carried out according to the Guide for the Care and Use of Laboratory Animals.
The animals were divided into six groups (five rats per group), as follows: group 1 (OTMCS-PEI-R13-h/DNA, w/w 10); group 2 (OTMCS-PEI-R13-h/DNA, w/w 30); group 3 (OTMCS-PEI-R13-l/DNA, w/w 10); group 4 (OTMCS-PEI-R13-l/DNA, w/w 30); group 5 (OTMCS-PEI); and group 6 (PEI 25 kDa/DNA, N/P 5) HeLa cells were injected subcutaneously into the mice to establish the tumor model. When the tumor diameter reached 10 mm, 250 μL of complex solution containing 30 μg of the pGL3-control reporter gene was injected via the tail vein. After 24 hours, the mice were euthanized by cervical dislocation, and the major tissues and tumors were collected and homogenized in cell lysis buffer. The cell lysate was then centrifuged. The relative light units were detected against protein concentration in the cell tissue extracts, which was measured with the Bioteke PP1001 protein assay kit.
Statistical analysis
The data were shown as the mean ± standard deviation. Statistically significant differences were determined using two-sample Student’s t-tests and analysis of variance, with P<0.05 taken as the level of statistical significance.
Results and discussion
Synthesis and characterization of OTMCS-PEI-R13
A new nonviral gene delivery vector, OTMCS-PEI-R13, was synthesized by linking PEI 2 kDa with OTMCS and then conjugating this complex with a bifunctional peptide, ie, R13. SMCC was used as a cross-linker to conjugate OTMCS-PEI with R13.
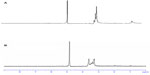
Figure 1A and B show the 1H NMR spectra for OTMCS-PEI and OTMCS-PEI-R13 in D2O, respectively. In Figure 1A, the proton peak at δ0.8–1.0 ppm represents −CH3, at δ2.9–3.3 ppm represents −CH2CH2O−, and at δ4.7–4.9 ppm represents −CH2CH2NH−. Compared with Figure 1A, the main absorption peaks for OTMCS-PEI-R13 in Figure 1B moved toward a lower magnetic field because of the strong electron shielding effect of R13; at the same time, the proton peaks at δ3.2–3.7 ppm were flattened and the peak value at δ1.00 ppm decreased due to the presence of carbonyl groups in R13. Further, at δ4.7–4.9 ppm, the peak value of the proton peak increased after introduction of R13. These results indicate that R13 had been coupled successfully to OTMCS-PEI.
Degradation studies
Once a nondegradable carrier has transferred pDNA into target cells, it cannot be degraded or eliminated from these cells. Such intracellular accumulation could potentially lead to cytotoxicity, so degradation of the gene vector is very important for gene transfection.
The ester bonds of OTMCS-PEI-R13 are susceptible to hydrolysis under physiological conditions, so the polymer can degrade into oligomers and low molecular weight PEI, which can be secreted into the extracellular environment by exocytosis, while having a limited toxic effect on cells. The average results for the in vitro degradation test are shown in Figure 2. We found that the original molecular weight of OTMCS-PEI-R13 determined by gel permeation chromatography was about 31,000. It was degraded slowly with the passage of time, and after about 60 hours, it was degraded almost completely as a result of hydrolysis of the ester bonds, and the final molecular weight was about 6,000.
To obtain meaningful information concerning the degradation model, the degradation profile was fitted to three different kinetic models, and the goodness of fit of the degradation data was assessed. A zero-order model was used to describe the degradation profile of OTMCS-PEI-R13. According to this model, the half-life of OTMCS-PEI-R13 was about 30 hours.
Buffering capacity of the OTMCS-PEI-R13 polymer
The buffering capacity of the gene vector is vital to the molecules escaping the endosomes of cells, because the molecules entering cells will experience a drop of pH from neutral to about 5. The proton sponge effect of the polymer ensures buffering in the endosomes, resulting in degradation of lysosomes so that the genes can be protected. Figure 3 shows that OTMCS-PEI-R13 had relatively higher buffering capacity in the pH range of 4–8 compared with H2O, suggesting that OTMCS-PEI-R13 is suitable for gene transfection.
Particle size, zeta potential measurement, and morphology
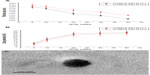
The particle size and zeta potential of the OTMCS-PEI-R13/DNA complexes were examined at different w/w ratios. As shown in Figure 4A, the particle size decreased when the weight ratio of OTMCS-PEI-R13/DNA increased, indicating that complexes bearing more charge could condense DNA more effectively. All the polymers tested could concentrate pDNA into nanoparticles, and depending on the composition of the complexes, their average particle size varied from 150 nm to 250 nm, and was suitable for efficient gene delivery in vivo. In the range of concentrations studied, no precipitation was observed at any w/w ratio.
The positively charged surfaces of the complexes are thought to be important for binding to negatively charged cellular membranes and cellular uptake via endocytosis.27 However, strong cationic charges can lead to cytotoxicity. The zeta potential is a measure of the overall surface charge of the nanoparticles. Figure 4B shows that the zeta potential of the complexes increased with an increase in their weight ratio. At low w/w ratios, the zeta potential of the complexes was slightly positive, and increased rapidly up to a w/w ratio of 20. Beyond this ratio, the increase became slow.
Figure 4C shows that the OTMCS-PEI-R13/DNA complex is a spherical structure with a particle size of about 100 nm, demonstrating that micelles were formed successfully.
Condensation of pDNA by OTMCS-PEI-R13
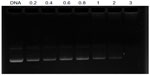
The DNA condensation capacity of OTMCS-PEI-R13 at different ratios was determined by agarose gel retardation assay. As the proportion of OTMCS-PEI-R13 increased, movement of pDNA became retarded. When the DNA was completely condensed by OTMCS-PEI-R13, it stopped migrating to the anode. As shown in Figure 5, OTMCS-PEI-R13 was able to condense DNA effectively and neutralized its charge at a w/w ratio of 3. When the w/w ratio exceeded 3, the complexes contained positive charges and therefore stopped migrating to the anode. Our previous work has demonstrated that, for unconjugated OTMCS-PEI, DNA was retarded at a w/w ratio of 0.6 (data not shown).22 Correspondingly, more OTMCS-PEI-R13 than OTMCS-PEI was needed to condense the pDNA completely, possibly because of more primary amines from OTMCS-PEI being substituted by R13 so that fewer positively charged primary amines interacted with the negatively charged pDNA phosphates.28
Protection of OTMCS-PEI-R13 on pDNA
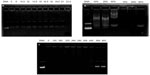
The OTMCS-PEI-R13 protected DNA from digestion when the complex solution was incubated with DNase I. As shown in Figure 6A, most of the DNA remained protected from digestion until the concentration of DNase I reached 23.5 U/μg DNA. In fact, under the same experimental conditions, DNA can be completely digested at a DNase I concentration of 0.08 U/μg DNA.29
The stability of the OTMCS-PEI-R13/DNA complex in vivo was evaluated by treating the complex with serum and sodium heparin in vitro. As shown in Figure 6B, when DNA was coated completely by OTMCS-PEI-R13, there were no electrophoretic bands of DNA, so serum could not dissociate the OTMCS-PEI-R13/DNA complex even at a concentration of 50%. Therefore, it could be inferred that the OTMCS-PEI-R13/DNA complex would remain stable in the circulation.
Sodium heparin can be used to simulate molecules carrying a negative charge in the blood, and can also dissociate gene vectors due to their negative charge. Sodium heparin can dissociate such complexes in a concentration-dependent manner.29 As shown in Figure 6C, at a w/w ratio of 20, sodium heparin could dissociate the OTMCS-PEI-R13/DNA complex at a concentration of 500 μg/mL. The complexes were completely dissociated when the concentration exceeded 600 μg/mL. These results indicate that OTMCS-PEI-R13/DNA complexes have a strong ability to resist sodium heparin.
Cytotoxicity assay
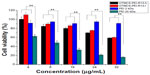
The ability of high molecular weight PEI to act as a transfection reagent is limited by its relatively high cytotoxicity, so we attempted to introduce a natural compound to form degradable PEI via linkers.30–32 In the present study, the cytotoxicity of OTMCS-PEI-R13 was evaluated in HeLa cells using the MTT assay and compared with PEI 25 kDa.
Figure 7 showed that PEI 25 kDa was more cytotoxic at a concentration of 8 μg/mL, where more than 50% of cells died, and cell viability fell to less than 20% at a concentration of 32 μg/mL. In contrast, OTMCS-PEI-R13 showed much higher cell viability than PEI 25 kDa at any concentration (P<0.01). More than 60% of cells survived under the influence of OTMCS-PEI-R13 at the highest concentration (32 μg/mL), indicating that OTMCS-PEI-R13 is suitable as a gene vector. The lower cytotoxicity of OTMCS-PEI-R13 may be due to its amino group density being lower than that of PEI 25 kDa. Further, the ester bonds in OTMCS-PEI-R13 could degrade into oligomers and low molecular weight PEI under physiological conditions, and could be rapidly eliminated from the cell, resulting in less cytotoxicity.
Transfection efficiency in vitro
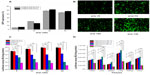
The in vitro transfection efficiency of OTMCS-PEI-R13 was investigated in HeLa cells using pEGFP-N2 and pGL3-control reporter genes. The OTMCS-PEI-R13/DNA complex was prepared at weight ratios of 5, 10, 20, and 30, respectively. PEI 25 kDa and PEI 2 kDa were prepared at the same weight ratios as controls.
Figure 8A shows GFP reporter gene expression in HeLa cells by flow cytometry analysis. Figure 8B shows the representative fluorescence images for the transfection of HeLa cells using OTMCS-PEI-R13 at weight ratios of 5, 10, 20, and 30 respectively. As shown in Figure 8A and B, the transfection efficiency of OTMCS-PEI-R13 complexes increased with the w/w ratios as indicated by bright green fluorescent segments showing that the pEGFP reporter gene was effectively transfected into HeLa cells.
Figure 8C shows the gene transfection efficiency of OTMCS-PEI-R13/pGL3-control complexes in HeLa cells compared with other controlled complexes. The OTMCS-PEI-R13 complexes showed much higher gene transfection than OTMCS-PEI, PEI 25 kDa, and PEI 2 kDa, and had maximal transfection efficiency at a w/w ratio of 30. The highest luciferase expression obtained was more than 60,000 times higher than for PEI 2 kDa, and approximately 7,000 times higher than for PEI 25 kDa at optimal conditions. PEI 25 kDa showed the best transfection efficiency at a w/w ratio of 10. As the w/w ratio increased, gene transfection decreased, mainly as a result of high cytotoxicity.
It should be noted that the R13 peptide could not only target the OTMCS-PEI-R13/DNA complexes to cells but could also promote cargo transport. As shown in Figure 8C, OTMCS-PEI-R13/DNA complexes showed a gene transfection efficiency that was about 41 times greater than that of the OTMCS-PEI/DNA complexes. In addition, when the R13 ratio increased, the transfection efficiency of OTMCS-PEI-R13 decreased, which can be explained by the high degree of modification of R13 leading to a less positive surface.
Transfection efficiency in vivo
pGL3-control gene transfection was performed in vivo using a tail vein injection of pDNA complexed with OTMCS-PEI-R13 to evaluate DNA delivery efficiency for OTMCS-PEI-R13. As shown in Figure 8D, the transfection efficiency of the OTMCS-PEI-R13 complexes was markedly higher than that of OTMCS-PEI and PEI 25 kDa in every tissue, and increased with w/w ratios, which is in accordance with the experimental results obtained in vitro and also indicates an increase in penetrating efficiency. Further, compared with OTMCS-PEI, the distribution of OTMCS-PEI-R13 in tumor tissue was markedly enhanced, indicating the tumor-targeting specificity of R13. pGL3-control reporter gene expression using the OTMCS-PEI-R13/DNA complexes was highest in the lung, followed by subcutaneous transplanted tumor and liver. This may be due to the fact that the larger complexes were easily entrapped mechanically because of the abundance of capillaries in the lung. In addition, the complexes with surface positive charges were easily taken up by the lung due to electrostatic interaction.33,34
The transfection efficiency assays in vitro and in vivo showed that these micelle-like structures based on amphiphilic chitosan, low molecular weight PEI, and the bifunctional peptide R13 had reduced cytotoxicity and enhanced transfection efficiency and tumor-targeting.
Conclusion
We developed a novel degradable OTMCS-PEI-R13 gene vector by cross-linking low molecular weight PEI with OTMCS and coupling this complex with a bifunctional peptide, R13. Using various physicochemical methods, we confirmed that OTMCS-PEI-R13 has the ability to form complexes with DNA and has suitable physicochemical properties for gene delivery.
The vector showed controlled degradation fitting a zero-order model. The half-life was about 30 hours, during which OTMCS-PEI-R13 could deliver DNA to the nucleus and be degraded into micromolecules. The polymer showed favorable buffering capacity. The particle size of the OTMCS-PEI-R13/DNA complexes was around 150–250 nm and the zeta potential ranged from 10 mV to 30 mV, which is appropriate for a gene vector. The OTMCS-PEI-R13 polymer was able to condense DNA completely at a weight ratio of 3. The polymer was able to protect pDNA from digestion at a DNase I concentration of 23.5 U/μg DNA. At the same time, OTMCS-PEI-R13 was able to resist dissociation induced by 50% fetal bovine serum and sodium heparin 400 μg/mL. Further, this novel gene vector had much lower cytotoxicity and higher gene transfection efficiency than PEI 25 kDa in HeLa cell lines. The transfection efficiency of the polymer in vivo was also higher than that of PEI 25 kDa. The cell selectivity and penetration ability of OTMCS-PEI-R13/DNA were also improved when compared with OTMCS-PEI, and consequently enhanced the transfection efficiency and tumor targeting specificity. In conclusion, OTMCS-PEI-R13 is a potential candidate for gene delivery, with low cytotoxicity and high transfection efficiency, as well as good tumor-targeting specificity.
Acknowledgment
This study was supported by the National Natural Science Foundation of China (81001024) and by the Scientific Research Innovation Project of the Shanghai Municipal Education Commission (13YZ097).
Disclosure
The authors report no conflicts of interest in this work.
References
Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29:3477–3496. | |
Danielsen S, Strand S, de Lange Davies C, Stokke BT. Glycosaminoglycan destabilization of DNA-chitosan polyplexes for gene delivery depends on chitosan chain length and GAG properties. Biochim Biophys Acta. 2005;1721:44–54. | |
Verma IM, Somia N. Gene therapy-promises, problems and prospects. Nature. 1997;389:239–242. | |
Peng M, Liu W, Yang G, et al. Investigation of the degradation mechanism of cross-linked polyethyleneimine by NMR spectroscopy. Polym Degrad Stab. 2008;93:476–482. | |
Guo Q, Shi S, Wang X, et al. Synthesis of a novel biodegradable poly(ester amine) (PEAs) copolymer based on low-molecular-weight polyethyleneimine for gene delivery. Int J Pharm. 2009;379:82–89. | |
Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjug Chem. 2001;12:989–994. | |
Remaut K, Sanders NN, Geest BG, Braeckmans K, Demeester J, De Smedt SC. Nucleic acid delivery: where material sciences and bio-sciences meet. Mater Sci Eng R Rep. 2007;58:117–161. | |
Seow WY, Yang YY. Functional polycarbonates and their self-assemblies as promising non-viral vectors. J Control Release. 2009;139:40–47. | |
Wu X, Ding B, Gao J, et al. Second-generation aptamer-conjugated PSMA-targeted delivery system for prostate cancer therapy. Int J Nanomedicine. 2011;6:1747–1756. | |
Weiss SI, Sieverling N, Niclasen M, et al. Uronic acids functionalized polyethyleneimine (PEI)-polyethyleneglycol (PEG)-graft-copolymers as novel synthetic gene carriers. Biomaterials. 2006;27:2302–2312. | |
Deng R, Yue Y, Jin F, et al. Revisit the complexation of PEI and DNA – how to make low cytotoxic and highly efficient PEI gene transfection non-viral vectors with a controllable chain length and structure? J Control Release. 2009;140:40–46. | |
Lungwitz U, Breunig M, Blunk T, Göpferich A. Polyethylenimine-based non-viral gene delivery systems. Eur J Pharm Biopharm. 2005;60:247–266. | |
Lin EH, Keramidas M, Rome C, et al. Lifelong reporter gene imaging in the lungs of mice following polyethyleneimine-mediated sleeping-beauty transposon delivery. Biomaterials. 2011;32:1978–1985. | |
Neu M, Fischer D, Kissel T. Recent advances in rational gene transfer vector design based on poly (ethylene imine) and its derivatives. J Gene Med. 2005;7:992–1009. | |
Huang H, Yu H, Tang G, Wang Q, Li J. Low molecular weight polyethylenimine cross-linked by 2-hydroxypropyl-gamma-cyclodextrin coupled to peptide targeting HER2 as a gene delivery vector. Biomaterials. 2010;31:1830–1838. | |
Kim TH, Cook SE, Arote RB, et al. A degradable hyperbranched poly (ester amine) based on poloxamer diacrylate and polyethylenimine as a gene carrier. Macromol Biosci. 2007;7:611–619. | |
Romøren K, Thu BJ, Evensen Ø. Immersion delivery of plasmid DNA. II. A study of the potentials of a chitosan based delivery system in rainbow trout (Oncorhynchus mykiss) fry. J Control Release. 2002;85:215–225. | |
Manuel AM, Taboada P, Sabín J, Krajewska B, Varela LM, Rodríguez JR. DNA-chitosan complexation: a dynamic light scattering study. Colloids Surf A Physicochem Eng Asp. 2009;339:145–152. | |
Turan K, Nagata K. Chitosan-DNA nanoparticles: the effect of cell type and hydrolysis of chitosan on in vitro DNA transfection. Pharm Dev Technol. 2006;11:503–512. | |
Gao Y, Zhang ZW, Chen LL, Gu WW, Li YP. Chitosan N-betainates/DNA self-assembly nanoparticles for gene delivery: in vitro uptake and transfection efficiency. Int J Pharm. 2009;371:156–162. | |
Lai WF, Lin MC. Nucleic acid delivery with chitosan and its derivatives. J Control Release. 2009;134:158–168. | |
Liu CC, Zhu Q, Wu WH, et al. Degradable copolymer based on amphiphilic N-octyl-N-quatenary chitosan and low-molecular weight polyethylenimine for gene delivery. Int J Nanomedicine. 2012;7:5339–5350. | |
Ogawa M, Hatano K, Oishi S, et al. Direct electrophilic radiofluorination of a cyclic RGD peptide for in vivo αvβ3 integrin related tumor imaging. Nucl Med Biol. 2003;30:1–9. | |
Lee DE, Hong YD, Choi KH, Lee S-Y, Park PH, Choi SJ. Preparation and evaluation of 99mTc-labeled cyclic arginine-glycine-aspartate(RGD) peptide for integrin targeting. Appl Radiat Isot. 2010;68:1896–1902. | |
Santos-Cuevas CL, Ferro-Flores G, Murphy CA, et al. Design, preparation, in vitro and in vivo evaluation of 99mTc-N2S2-Tat(49–57)-bombesin: a target-specific hybrid radiopharmaceutical. Int J Pharm. 2009;375:75–83. | |
Liu KH, Wang XY, Fan W, et al. Degradable polyethylenimine derivate coupled to a bifunctional peptide R13 as a new gene-delivery vector. Int J Nanomedicine. 2012;7:1149–1162. | |
Kim T-I, Baek J-U, Yoon JK, Choi JS, Kim K, Park J-S. Synthesis and characterization of a novel arginine-grafted dendritic block copolymer for gene delivery and study of its cellular uptake pathway leading to transfection. Bioconjug Chem. 2007;18:309–317. | |
Ma K, Hu MX, Qi Y, et al. PAMAM-triamcinolone acetonide conjugate as a nucleus-targeting gene carrier for enhanced transfer activity. Biomaterials. 2009;30:6109–6118. | |
Hao JG, Sha XY, Tang YJ, et al. Enhanced transfection of polyplexes based on pluronic-polypropylenimine dendrimer for gene transfer. Arch Pharm Res. 2009;32:1045–1054. | |
Anderson DG, Akinc A, Hossain N, Langer R. Structure/property studies of polymeric gene delivery using a library of poly(b-amino esters). Mol Ther. 2005;11:426–434. | |
Lin C, Blaauboer CJ, Timoneda MM, et al. Engbersen, bioreducible poly(amido amine)s with oligoamine side chains: synthesis, characterization, and structural effects on gene delivery. J Control Release. 2008;126:166–174. | |
Park MR, Han KO, Cho MH, Nah JW, Choi YJ, Cho CS. Degradable polyethylenimine-alt-poly (ethylene glycol) copolymers as novel gene carriers. J Control Release. 2005;105:367–380. | |
Kellaway IW, Farr SJ. Liposomes as drug delivery systems to the lung. Adv Drug Deliv Rev. 1990;5:149–161. | |
Kawakami S, Ito Y, Charoensit P, Yamashita F, Hashida M. Evaluation of proinflammatory cytokine production induced by linear and branched polyethylenimine/plasmid DNA complexes in mice. J Pharmacol Exp Ther. 2006;317:1382–1390. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.