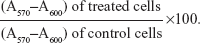Back to Journals » International Journal of Nanomedicine » Volume 9 » Issue 1
Increased gene delivery efficiency and specificity of a lipid-based nanosystem incorporating a glycolipid
Authors Magalhães M, Farinha D, Pedroso de Lima M, Faneca H
Received 23 June 2014
Accepted for publication 27 August 2014
Published 24 October 2014 Volume 2014:9(1) Pages 4979—4989
DOI https://doi.org/10.2147/IJN.S69822
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Mariana Magalhães,1,2,* Dina Farinha,1,* Maria Conceição Pedroso de Lima,1,2 Henrique Faneca1
1Center for Neuroscience and Cell Biology, University of Coimbra, Coimbra, Portugal; 2Department of Life Sciences, Faculty of Science and Technology, University of Coimbra, Coimbra, Portugal
*These authors contributed equally to this work
Abstract: Hepatocellular carcinoma (HCC) is the third most common cause of death related to cancer diseases worldwide. The current treatment options have many limitations and reduced success rates. In this regard, advances in gene therapy have shown promising results in novel therapeutic strategies. However, the success of gene therapy depends on the efficient and specific delivery of genetic material into target cells. In this regard, the main goal of this work was to develop a new lipid-based nanosystem formulation containing the lipid lactosyl-PE for specific and efficient gene delivery into HCC cells. The obtained results showed that incorporation of 15% of lactosyl-PE into liposomes induces a strong potentiation of lipoplex biological activity in HepG2 cells, not only in terms of transgene expression levels but also in terms of percentage of transfected cells. In the presence of galactose, which competes with lactosyl-PE for the binding to the asialoglycoprotein receptor (ASGP-R), a significant reduction in biological activity was observed, showing that the potentiation of transfection induced by the presence of lactosyl-PE could be due to its specific interaction with ASGP-R, which is overexpressed in HCC. In addition, it was found that the incorporation of lactosyl-PE in the nanosystems promotes an increase in their cell binding and uptake. Regarding the physicochemical properties of lipoplexes, the presence of lactosyl-PE resulted in a significant increase in DNA protection and in a substantial decrease in their mean diameter and zeta potential, conferring them suitable characteristics for in vivo application. Overall, the results obtained in this study suggest that the potentiation of the biological activity induced by the presence of lactosyl-PE is due to its specific binding to the ASGP-R, showing that this novel formulation could constitute a new gene delivery nanosystem for application in therapeutic strategies in HCC.
Keywords: targeted gene delivery, hepatocellular carcinoma, lactosyl-PE, cationic liposomes, gene delivery nanosystems
Introduction
Hepatocellular carcinoma (HCC) is one of the primary hepatic neoplasms of liver cancer1,2 and is considered the sixth most common cancer type with the highest prevalence worldwide, corresponding to the third most common cause of death related to cancer diseases.3,4 This high mortality is due to the fact that HCC is only detected at advanced stages of the disease (the early stages are asymptomatic) and the current treatment options available, which consist of tumor surgical recession, liver transplantation, and chemotherapy, are limited and have a reduced application rate.5–7 In this context, the development of efficient and specific antitumor strategies for HCC is urgently required, with gene therapy being considered a promising strategy to treat this disease.8
Gene therapy has been developed over the past years and is intended to use genetic material to prevent or treat monogenic diseases and acquired genetic pathologies, like cancer. However, it still has a limited clinical application, mainly due to the reduced gene delivery efficiency and specificity into target cells. For that reason, several types of gene delivery nanosystems have been investigated in order to achieve successful and efficient nucleic acid delivery into target cells and consequently the desired therapeutic effect.9–11 Among these, cationic liposome/DNA complexes (lipoplexes) have been the most extensively studied, since they present higher gene delivery efficiency, both in vitro and in vivo, than that observed with other non-viral gene delivery systems.12,13,14 Our studies have demonstrated that cationic liposomes composed of 1-palmitoyl-2-oleoyl-sn-glycero-3-ethylphosphocholine (EPOPC) and cholesterol (Chol), at a molar ratio of 1:1, have the ability to mediate efficient gene delivery in different tissues and cells lines.13,15 Lipoplexes prepared with these cationic liposomes were shown to exhibit higher levels of transfection than those obtained with nanosystems prepared with other cationic liposome formulations.15 In addition, EPOPC, which is a derivative of a natural lipid (phosphatidylcholine), has revealed lower cytotoxicity due the presence of ester linkages and, therefore is suitable for clinical applications of gene therapy.13,15–17
Liver has several characteristics that make it very attractive for application of gene therapy, namely the presence of the asialoglycoprotein receptor (ASGP-R). ASGP-R is an integral membrane protein specifically expressed in hepatocytes and overexpressed in tumoral hepatocytes,18–20 allowing the development of therapies targeted to these cells. In normal hepatocytes, this receptor is distributed in a polarized way, being located at the basolateral zone,18 whereas in tumoral hepatocytes this overexpressed receptor loses its polarized distribution.20 This receptor specifically binds terminal residues of galactose or N-acetylgalactosamine present on desialylated glycoproteins or glycolipids.21 In this context, our rational was that the incorporation of a glycolipid, containing a galactose terminal residue, into the lipid-based nanosystem could promote its targeting to HCC cells and cellular internalization through receptor-mediated endocytosis, consequently leading to a substantial increase of gene delivery into these cells.
The main goal of this work was to develop a new gene delivery nanosystem, prepared with EPOPC:Chol:lactosyl-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-lactosyl [ammonium salt]) cationic liposomes, and to evaluate its efficacy and specificity to deliver genetic material into HCC cells, as well as to study the possible mechanisms by which this nanosystem mediates biological activity, and to analyze its physicochemical properties.
Materials and methods
Cell culture
HepG2 cells (human HCC) and MDA-MB-231 cells (human breast adenocarcinoma) were maintained at 37°C, 5% CO2, in Dulbecco’s Modified Eagle’s Medium-high glucose (DMEM-HG) (Sigma-Aldrich Co., St Louis, MO, USA) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Sigma-Aldrich Co.), penicillin (100 U/mL) and streptomycin (100 μg/mL).
Preparation of cationic liposomes and lipoplexes
Small unilamellar cationic liposomes were prepared by extrusion of multilamellar liposomes composed of 1:1 molar ratio mixtures of EPOPC and Chol, and different amounts of lactosyl-PE. EPOPC:Chol:lactosyl-PE liposomes were labeled with 1% rhodamine-dioleoyphosphatidylethanolamine (rhodamine-PE), for cell uptake studies. Briefly, lipids (Avanti Polar Lipids, Alabaster, AL, USA) dissolved in CHCl3 were mixed at the desired molar ratio and dried under vacuum in a rotator evaporator. The dried lipid films were hydrated with deionized water to a final lipid concentration of 6 mM and the resulting multilamellar liposomes were then sonicated for 3 minutes and extruded 21 times through two stacked polycarbonate filters of 50 nm pore diameter using a Liposofast device (Avestin, Toronto, ON, Canada). The resulting liposomes were then diluted four times with deionized water and filter-sterilized utilizing 0.22 μm pore-diameter filters (Schleicher & Schuell, Dassel, Germany). The suspension was stored at 4°C until use. Lipoplexes were prepared by sequentially mixing 100 μL of a HEPES-buffered saline solution (HBS) (100 mM NaCl, 20 mM HEPES, pH 7.4), with liposomes (the volume necessary to obtain the desired [+/−] lipid/DNA charge ratio) and with 100 μL of HBS solution containing 1 μg of pCMVluc-encoding luciferase (a gift of Dr P Felgner, Vical, San Diego, CA, USA), or 1 μg of pCMVgfp-encoding green fluorescent protein (GFP) (Clontech Laboratories, Mountain View, CA, USA). The mixtures were further incubated for 15 minutes at room temperature. Lipoplexes were used immediately after being prepared.
Transfection activity
For luminescence evaluation of luciferase expression, 80×103 HepG2 cells/well or 75×103 MDA-MB-231 cells/well were seeded in 48-well culture plates 24 hours before transfection. Cells were used at 70%–80% confluence, and lipoplexes containing 1 μg of pCMVluc plasmid DNA were added to cells, previously covered with 0.3 mL of DMEM-HG. After 4 hours’ incubation (5% CO2 at 37°C), the transfection medium was replaced with DMEM-HG and cells were further incubated for 48 hours. Cells were then washed twice with phosphate-buffered saline solution (PBS) and 150 μL of lysis buffer (composed of 1 mM dithiothreitol; 1 mM ethylenediaminetetraacetic acid; 25 mM Tris (1,3-dichloro-2-propyl)-phosphate [pH 7.8]; 8 mM MgCl2; 15% glycerol; and 1% [v/v] Triton X-100™), and were added to each well. The levels of luciferase expression in lysates were quantified by measuring light production by luciferase in an Lmax II 384 luminometer (Molecular Devices, Sunnyvale, CA, USA). The protein content of the lysates was measured by the DC™ Protein Assay reagent (Bio-Rad Laboratories Inc., Hercules, CA, USA) using bovine serum albumin as the standard. The data were expressed as relative light units of luciferase per milligram of total cell protein.
For flow cytometry analysis of GFP expression, 320×103 HepG2 cells/well were seeded in 12-well culture plates and, after 24 hours, lipoplexes containing 4 μg of pCMVgfp were added to cells. After 4 hours’ incubation (5% CO2 at 37°C), the transfection medium was replaced with DMEM-HG, and cells were further incubated for 48 hours. Cells were then washed twice with PBS and detached with trypsin (5 minutes at 37°C). Thereafter, cells were washed and resuspended in PBS, and immediately analyzed in a FACSCalibur™ flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Live cells were gated by forward/side scattering from a total of 25,000 events, and data were analyzed using CellQuest™ software.
For fluorescence microscopy analysis of GFP expression, 320×103 HepG2 cells/well were seeded in 12-well culture plates (previously covered with a coverslip) and after 24 hours lipoplexes containing 4 μg of pCMVgfp were added to the cells. After 4 hours’ incubation (5% CO2 at 37°C), the transfection medium was replaced with DMEM-HG, and cells were further incubated for 48 hours. Cells were then washed twice with PBS, fixed with 4% paraformaldehyde for 15 minutes at room temperature, and then mounted in MowiolR mounting medium (Sigma-Aldrich Co.). The Images (original magnification: ×20) were obtained on an Axioskop 2 Plus microscope (Zeiss, Munich, Germany) using an AxioCam HRc camera (Zeiss).
For the competitive studies, the culture medium containing different concentrations of galactose (20, 40, 80, and 100 mg/mL) were added to cells 30 minutes before the addition of the nanosystems and maintained during the 4 hours of transfection. In the biological activity studies using an antibody against the ASGP-R (Santa Cruz Biotechnology Inc., Dallas, TX, USA), the culture medium containing 40 μg/mL of antibody was added to HepG2 cells 30 minutes before the lipoplexes’ addition and maintained during the 4 hours of transfection.
Cell viability
Following transfection under the different experimental conditions, cell viability was assessed by a modified Alamar® Blue assay. This assay measures the redox capacity of the cells due to the production of metabolites as a result of cell growth. Briefly, 0.3 mL of 10% (v/v) Alamar Blue dye in complete DMEM-HG medium, prepared from a 0.1 mg/mL stock solution of Alamar Blue, was added to each well 47 hours following the initial period of transfection (4 hours). After 1 hour of incubation at 37°C, 170 μL of the supernatant were collected from each well and transferred to 96-well plates. The absorbance at 570 and 600 nm was measured in a SPECTRAmax® PLUS 384 spectrophotometer (Molecular Devices). Cell viability (as a percentage of untreated control cells) was calculated according to the formula:
| (1) |
Cell binding and uptake
EPOPC:Chol (1:1) liposomes or EPOPC:Chol:lactosyl-PE (15%) liposomes, labeled with 1% rhodamine-PE, were complexed with 1 μg of pCMVluc at 4/1 and 2/1 (+/−) cationic lipid/DNA charge ratios. Twenty-four hours before transfection, HepG2 cells were seeded in 48-well culture plates. Thirty minutes before transfection, cells were incubated with 0.3 mL of DMEM-HG medium, either containing or not containing 40 mg/mL of galactose, and the lipoplexes were then added to each well. Following 4 hours’ incubation at either 4°C or 37°C, cells were washed twice with PBS and lysed with 100 μL/well of 1% Triton X-100. Binding (4°C) and uptake (37°C) of lipoplexes were monitored in a SPECTRAmax GEMINI™ EM fluorometer (Molecular Devices) by measuring the fluorescence at excitation and emission wavelengths of 545 and 587 nm, respectively.
Zeta potential
Nanosystems were characterized with respect to their zeta potential using a Zetasizer Nano (Malvern Instruments, Malvern, UK) that measures the electrophoretic mobility and zeta potential distribution by a phase analysis light scattering method. The analysis was performed at 25°C in HBS, and the lipoplexes were prepared immediately before analysis.
Transmission electron microscopy
Nanosystems were analyzed in terms of their size/structure by transmission electron microscopy (TEM). The prepared lipoplexes were absorbed on copper grids covered with Formvar® film for 2 minutes and then dried until touch-dry with filter paper. To enhance contrast, the grids were put on a drop of 0.5% uranyl acetate for 15 seconds. Excess stain was removed by filter paper. The grids were examined in a JEM 1400 transmission electron microscope (JEOL, Tokyo, Japan) at 80 kV accelerating voltage. Digital images were digitally recorded using a SC1000 ORIUS™ CCD camera (Gatan Inc., Warrendale, PA, USA).
Ethidium bromide intercalation
The accessibility of ethidium bromide to the DNA of lipoplexes was analyzed by fluorescence measurements in a SPECTRAmax GEMINI EM fluorometer (Molecular Devices). One hundred microliters of nanosystems, containing 0.5 μg of DNA, were transferred into a black 96-well plate (Sigma-Aldrich Co.), and then 100 μL of EtBr solution was added to each well to achieve a final EtBr concentration of 400 nM. Following 10 minutes’ incubation, fluorescence was measured with excitation and emission wavelengths of 518 and 605 nm, respectively. The fluorescence scale was calibrated such that the initial fluorescence of EtBr (100 μL of EtBr solution added to 100 μL of HBS) was set at residual fluorescence. The value of fluorescence obtained with 0.5 μg of naked DNA (control) was set as 100%. The amount of DNA available to interact with the probe was calculated by subtracting the values of residual fluorescence from those obtained for the sample and expressed as the percentage of the control.
Resistance to deoxyribonuclease I action
Resistance of nanosystems to deoxyribonuclease I (DNase I; Sigma-Aldrich Co.), which was maintained in an appropriate buffer (50 mM Tris-HCl [pH 7.5], 10 mM MnCl2, 50 μg/mL BSA), was determined by electrophoresis. Lipoplexes were submitted to DNase I action (2 units of DNase I/μg of DNA) for 20 minutes at 37°C, followed by inactivation of the enzyme upon incubation with 0.5 M EDTA (1 μL/unit of DNase I). Parallel experiments were performed by incubating samples under the same experimental conditions, with DNase I previously inactivated. Electrophoresis was performed in 1% agarose gel prepared in TBE solution (89 mM Tris-buffer [pH 8.6], 89 mM boric acid, 2.5 mM EDTA) and containing 1 μg/mL of EtBr. Following incubation of lipoplexes with DNase I (active or inactive), aliquots corresponding to 0.5 μg of DNA to which 5 μL of loading buffer (15% [v/v] Ficoll® PM 400; 0.05% [w/v] bromophenol blue; 1% [w/v] SDS; and 0.1 M EDTA, pH 7.8) had been previously added, were placed in the gel. The electrophoresis elapsed for 1 hour, applying a voltage of 80 V.
Statistical analysis
Data were analyzed using the Prism software (version 5.0). Statistical significance of differences between data was evaluated by one-way analysis of variance using the Tukey test. A value of P<0.05 was considered significant.
Results
Effect of lactosyl-PE on the biological activity and cytotoxicity of the lipoplexes in HepG2 cells
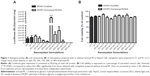
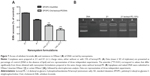
Since the ASGP-R is overexpressed in HCC cells, such as the HepG2 cell line, and specifically binds galactose, we evaluated what effect incorporating lactosyl-PE (which has a galactose terminal residue) into the cationic liposomes has on the biological activity of the generated nanosystem in HepG2 cells. For this purpose, we prepared several EPOPC:Chol cationic liposome formulations containing different amounts of lactosyl-PE (5%, 10%, 15%, 20%, and 40%) in order to verify how this lipid affects the biological activity of lipoplexes prepared at 2/1 and 4/1 (+/−) charge ratios. As illustrated in Figure 1A, the incorporation of lactosyl-PE into the EPOPC:Chol cationic liposomes could significantly increase the biological activity of the generated nanosystems, this being particularly evident for EPOPC:Chol:lactosyl-PE/DNA lipoplexes prepared with cationic liposomes containing 15% of lactosyl-PE at the 2/1 (+/−) charge ratio. For this formulation, a 50-fold increase in the transfection activity was observed when compared to that obtained with plain lipoplexes prepared at the same charge ratio with EPOPC:Chol cationic liposomes without lactosyl-PE. This level of potentiation was not verified for any of the other tested formulations. The effect of lactosyl-PE incorporation was evaluated only for lipoplexes prepared at 2/1 and 4/1 (+/-) charge ratios, since these formulations were the ones that presented the highest levels of biological activity (data not shown). On the other hand, results illustrated in Figure 1B show that the viability of HepG2 cells was not significantly affected (P>0.05) by any of the tested lipoplex formulations.
Effect of the presence of lactosyl-PE on the percentage of transfected HepG2 cells
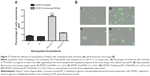
In order to assess the transfection efficiency of the generated nanosystems, which shows the amount of cells that could directly benefit from a gene therapy strategy, we prepared lipoplex formulations with the plasmid DNA encoding the GFP and analyzed the percentage of transfected cells by flow cytometry and fluorescence microscopy (Figure 2). The results obtained by flow cytometry (Figure 2A) showed that the percentage of transfected cells obtained with the best formulation, EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1 lipoplexes, was approximately 40%, this being much higher than that observed with plain lipoplexes (without lactosyl-PE) prepared at the same charge ratio (EPOPC:Chol/DNA [+/-] 2/1). On the other hand, for lipoplexes prepared at the 4/1 (+/-) charge ratio, the incorporation of 15% of lactosyl-PE promoted only a slight increase in the percentage of transfected cells.
These flow cytometry data were confirmed by fluorescent microscopy analysis of transfected HepG2 cells. As illustrated in Figure 2B, transfection performed with EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1 lipoplexes (panel III) resulted in a much higher number of GFP-expressing cells than that obtained with lipoplexes prepared at the same (+/-) charge ratio without lactosyl-PE (panel I). Moreover, it was observed that EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 4/1 lipoplexes (panel IV) were much less efficient than the best formulation (panel III), which is consistent with the results obtained by flow cytometry (Figure 2A).
Biological activity of the nanosystems containing lactosyl-PE in ASGP-R-non-expressing cells and in the presence of galactose or an antibody against the ASGP-R
In order to verify whether the potentiation of the biological activity of lipoplexes, induced by the incorporation of lactosyl-PE, was due to the specific interaction of its galactose terminal residue with ASGP-R, we evaluated the transfection activity of the generated nanosystems in MDA-MB-231 cells, that do not express the ASGP receptor, and in HepG2 cells both in the presence of galactose, which acts as a competitive agent to the binding to ASGP-R, and in the presence of an antibody to block the ASGP receptor.
As opposed to what was observed with HepG2 cells (Figure 1A), the results presented in Figure 3 showed no increase in the biological activity of lipoplexes prepared with lactosyl-PE in MDA-MB-231 cells, when compared to that obtained with the corresponding plain lipoplexes for both charge ratios. On the other hand, as illustrated in Figure 4A, the results obtained in the transfection assays performed in culture medium containing 40 mg/mL of galactose revealed that the presence of free galactose promoted a strong reduction in the biological activity of the best formulation (EPOPC:Chol:lactosyl-PE [15%]/DNA [+/-] 2/1). In contrast, the biological activity of lipoplexes prepared without lactosyl-PE was not inhibited by the presence of 40 mg/mL of galactose (Figure 4A). Although several concentrations of galactose were tested in the transfection studies, the concentration of 40 mg/mL of galactose was selected for these studies since it was found to be the concentration at which a maximum inhibition of biological activity occurred without affecting cell viability (data not shown). Moreover, transfection studies performed in HepG2 cells treated with an antibody against ASGP-R showed that its presence induced a substantial inhibition of the biological activity of the EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1 lipoplexes, but not the transfection activity of the corresponding plain lipoplexes (Figure 4B).
Effect of lactosyl-PE and galactose on the cell binding and uptake of the nanosystems
In order to determine if the potentiation of the biological activity of the nanosystems, induced by the incorporation of lactosyl-PE, was due to the interaction of the galactose terminal residue of this glycolipid with ASGP-R, we evaluated the extent of cell binding and uptake of the generated formulations in HepG2 cells, in the presence or absence of galactose (40 mg/mL). For this purpose, cells were incubated with rhodamine-PE-labeled lipoplexes at 4°C (binding) or at 37°C (uptake). As illustrated in Figure 5, cell incubation with the best formulation (EPOPC:Chol:lactosyl-PE [15%]/DNA [+/-] 2/1) resulted in a much higher extent of cell binding and uptake than that obtained with the corresponding plain lipoplexes. This enhancing effect on cell binding and uptake promoted by lactosyl-PE was not observed for lipoplexes prepared at the 4/1 (+/-) charge ratio.
On the other hand, the presence of galactose induced a significant decrease in cell binding (P<0.01) and uptake (P<0001) of the EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1 lipoplexes, whereas no significant effect on these processes was observed for plain lipoplexes or those prepared with lactosyl-PE at the 4/1 (+/-) charge ratio.
Influence of lactosyl-PE on the physicochemical properties of the nanosystems
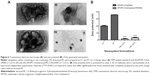
Since the efficacy of the nanocarriers to deliver genetic material into target cells is strongly dependent on their physicochemical properties, we determined their size/structure by TEM and surface charge by zeta potential. TEM analysis (Figure 6A) demonstrated that the lipoplexes prepared at 2/1 (+/-) charge ratio and containing 15% of lactosyl-PE (panels III and IV) presented considerably reduced mean diameters, approximately 200 nm, when compared to those obtained for the corresponding plain lipoplexes (panels I and II), approximately 800 nm. Moreover, TEM analysis showed that lipoplexes containing 15% of lactosyl-PE consisted of clusters of small nanostructures with sizes around 50 nm (panel III) that formed the observed 200 nm nanosystems (panels III and IV). The results obtained from zeta potential measurements (Figure 6B) showed that the nanosystems containing 15% of lactosyl-PE prepared at both 2/1 and 4/1 (+/-) charge ratios exhibited a positive zeta potential that is much lower than that observed for the corresponding plain lipoplex formulations.
Access to DNA associated to the nanosystems
In order to evaluate the capacity of the generated lipid-based nanosystems to condense and protect the carried DNA, we used the EtBr and DNase I assays. As illustrated in Figure 7A, nanosystems containing 15% of lactosyl-PE allowed much lesser access of EtBr to the carried DNA than that observed for the corresponding plain lipoplexes, at both 2/1 and 4/1 (+/-) charge ratios. The electrophoretic profile of lipoplexes submitted to DNase I assay (Figure 7B) showed that the intensity of the bands of EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1 lipoplexes, incubated with inactive DNase I, was smaller than that observed with the corresponding plain lipoplexes (submitted to the same experimental conditions), showing a lesser access of EtBr to DNA, in agreement to what was observed in the EtBr assay (Figure 7A). Moreover, EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1 lipoplexes exhibited a smaller difference between the intensity of their bands (incubation with inactive DNase I versus active DNase I), than the corresponding plain lipoplexes, showing that this new nanosystem confers a higher DNA protection.
Discussion
The development of specific and efficient gene delivery nanosystems for application in HCC will constitute a new platform for gene-based therapies for this disease, allowing the generation of more effective antitumor gene therapy strategies, such as those involving the delivery of the p53 gene, which is highly altered in this pathology, and those involving antiangiogenic approaches, for example against VEGF (vascular endothelial growth factor), due to the high vascularization of this type of tumor.22,23
ASGP-R is specifically expressed in hepatocytes and overexpressed in HCC cells, specifically recognizing galactose terminal residues, which could allow the development of targeted gene delivery systems for HCC. In this context, the use of the glycolipid lactosyl-PE, which contains a galactose residue in its polar group, can promote the nanosystem interaction with the ASGP-R and consequently induce its cellular internalization by endocytosis.21,24–27 On the other hand, we have previously shown that the use of cationic liposomes composed of EPOPC and Chol, at a 1:1 molar ratio, results in considerably higher biological activities than those obtained with other lipid-based formulations.13–17,28 In this regard, the main goal of this work consisted of the development of a new gene delivery lipid-based nanosystem for HCC, targeting the ASGP-R, based on cationic liposomes composed of EPOPC, Chol and lactosyl-PE.
We generated novel nanosystems (EPOPC:Chol:lactosyl-PE/DNA) and performed an extensive characterization in terms of biological activity and physicochemical properties, comparing them with formulations (EPOPC:Chol/DNA) previously developed in our laboratory.15 Our results demonstrated that the incorporation of lactosyl-PE in the nanocarriers (EPOPC:Chol:lactosyl-PE/DNA) promoted a substantial increase in their biological activity in HepG2 cells (Figures 1 and 2). This enhancing effect was dependent on the amount of lactosyl-PE and on the (+/-) charge ratio of lipoplexes, the highest increase in biological activity being achieved for nanosystems containing 15% of lactosyl-PE and prepared at the 2/1 (+/-) charge ratio. In fact, EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1 lipoplexes exhibited a much higher biological activity than the other developed formulations, which might be explained by a much better distribution of lactosyl-PE on its surface (due to a combination of several factors such as amount of lactosyl-PE, charge ratio, and structure of lipoplexes), promoting a higher interaction with ASGP-R, and also by a more favorable lipoplex structure, allowing a more efficient gene release into target cells. These different levels of biological activity obtained for the various formulations could not be associated with their cytotoxicity, since no significant reduction in the viability of HepG2 cells was observed after transfection with any of the tested formulations (Figure 1B). This potentiation was most probably due to interaction of the galactose residue of lactosyl-PE with ASGP-R, thereby enhancing cell binding and internalization of this nanosystem. In fact, the incorporation of lactosyl-PE into EPOPC:Chol/DNA (+/-) 2/1 lipoplexes resulted in a strong increase in their binding and uptake in HepG2 cells, as illustrated in Figure 5. On the other hand, the results obtained in MDA-MB-231 cells, which do not express ASGP-R, showed that the incorporation of 15% of lactosyl-PE into EPOPC:Chol/DNA lipoplexes did not promote any enhancement of transfection activity (Figure 3). These data also support the hypothesis that this new nanosystem (EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1) is specifically recognized by ASGP-Rs present on the surface of HepG2 cells, justifying the observed increase of biological activity (Figures 1A and 2). In addition, the results obtained in the competitive inhibition studies showed a strong decrease in the cell binding and uptake (Figure 5) of EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1 lipoplexes as well as in their biological activity (Figure 4A) in the presence of galactose, which is most probably due to competition of free galactose with the galactose terminal residue of lactosyl-PE for the binding to ASGP-R. Furthermore, the transfection activity of this new nanosystem was substantially inhibited by the presence of an antibody against ASGP-R (Figure 4B). These results demonstrate that the developed lipid-based nanosystem has the ability to specifically bind to ASGP-Rs of HCC cells through the galactose residues present in lactosyl-PE, promoting a high increase in their cellular internalization, and consequently, in their biological activity.
The efficacy of nanosystems to mediate gene delivery is greatly dependent on their physicochemical properties, since these are known to affect their stability and biological activity.12,29 TEM analysis (Figure 6A) showed that the incorporation of lactosyl-PE into EPOPC:Chol/DNA lipoplexes promoted a strong reduction in their mean diameter, which was most likely due to the formation of a layer of water over the nanosystems, induced by lactosyl-PE, that inhibited their aggregation, and thus led to smaller sizes. On the other hand, zeta potential measurements (Figure 6B) revealed that the incorporation of lactosyl-PE into EPOPC:Chol/DNA lipoplexes significantly reduced their positive net charge, which could be due to the ability of this glycolipid to mask the positive charge of the cationic lipid. Although EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1 nanosystems presented the lowest surface charge, and therefore less ability to establish electrostatic interactions with the negatively charged cell membrane, they exhibited the highest levels of cell binding, demonstrating that their binding was not due to non-specific electrostatic interactions but most likely to their specific interaction with ASGP-R. These data reinforce the hypothesis that this new nanosystem formulation specifically interacts with ASGP-R, which consequently promotes the uptake of these nanosystems by receptor-mediated endocytosis in HCC cells. Although lipoplexes containing lactosyl-PE presented a lower surface charge than the corresponding plain lipoplexes (Figure 6B), these new nanocarriers exhibited a greater ability to condense and protect the genetic material, as shown by the lesser access of EtBr to DNA and by the smaller DNA degradation induced by DNase I (Figure 7). This finding was most probably due to the presence of the glycolipid lactosyl-PE and to its ability to create a shield coating in the nanosystems, leading to a higher protection of the genetic material, thus ensuring the integrity of the delivered nucleic acids.17
Conclusion
In this work, we developed a new nanosystem formulation, EPOPC:Chol:lactosyl-PE (15%)/DNA (+/-) 2/1, that is easy to prepare and is easy to be submitted to scale transposition, which presents a high gene delivery efficiency and specificity to HCC cells, most probably attributed to its specific binding to ASGP-R. Moreover, our results show that this novel formulation exhibits adequate properties for in vivo application, namely absence of toxicity, high DNA protection, reduced surface charge, and low mean diameters.
Overall, our findings show that this new lipid-based nanosystem could be very useful for the development of efficient and specific antitumor gene therapy strategies towards application in HCC.
Acknowledgments
This work was financed by the grants PTDC/QUI-BIQ/116080/2009, PTDC/SAU-BMA/114482/2009 and PEst-C/SAU/LA0001/2013-2014 from the Portuguese Foundation for Science and Technology (FCT) and the European Community Fund (FEDER) through the COMPETE program.
Disclosure
The authors report no conflicts of interest in this work.
References
Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6(9):674–687. | ||
Li Y, Tang ZY, Hou JX. Hepatocellular carcinoma: insight from animal models. Nat Rev Gastroenterol Hepatol. 2012;9(1):32–43. | ||
Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. | ||
Gomaa AI, Khan SA, Toledano MB, Waked I, Taylor-Robinson SD. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World J Gastroenterol. 2008;14(270):4300–4308. | ||
Zhao X, Elmore LW, Harris CC, Wang XW. Hepatocellular carcinoma. In: eLS: Genetics and Disease Hoboken, NJ: John Wiley & Sons, Ltd.; 2011:1–7. | ||
Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53(3):1020–1022. | ||
Blum HE. Hepatocellular carcinoma: therapy and prevention. World J Gastroenterol. 2005;11(47):7391–7400. | ||
Sangro B, Mazzolini G, Ruiz M, et al. A phase I clinical trial of thymidine kinase-based gene therapy in advanced hepatocellular carcinoma. Cancer Gene Ther. 2010;17(12):837–843. | ||
Park J, Singha K, Son S, et al. A review of RGD-functionalized nonviral gene delivery vectors for cancer therapy. Cancer Gene Ther. 2012;19(11):741–748. | ||
Ginn SL, Alexander IE, Edelstein ML, Abedi MR, Wixon J. Gene therapy clinical trials worldwide to 2012 – an update. J Gene Med. 2013;15(2):65–77. | ||
Sheridan C. Gene therapy finds its niche. Nat Biotechnol. 2011;29(2):121–128. | ||
Faneca H, Simões S, Pedroso de Lima MC. Evaluation of lipid-based reagents to mediate intracellular gene delivery. Biochim Biophys Acta. 2002;1567(1–2):23–33. | ||
Simões S, Filipe A, Faneca H, et al. Cationic liposomes for gene delivery. Expert Opin Drug Del. 2005;2(2):237–254. | ||
Paecharoenchai O, Niyomtham N, Apirakaramwong A, et al. Structure relationship of cationic lipids on gene transfection mediated by cationic liposomes. AAPS Pharm Sci Tech. 2012;13(4):1302–1308. | ||
Faneca H, Faustino A, Pedroso de Lima MC. Synergistic antitumoral effect of vinblastine and HSV-Tk/GCV gene therapy mediated by albumin-associated cationic liposomes. J Control Release. 2008; 126(2):175–184. | ||
Pedroso de Lima MC, Simões S, Pires P, Faneca H, Düzgüneş N. Cationic lipid-DNA complexes in gene delivery: from biophysics to biological applications. Adv Drug Deliv Rev. 2001;47(2–3):277–294. | ||
Faneca H, Simões S, Pedroso de Lima MC. Association of albumin or protamine to lipoplexes: enhancement of transfection and resistance to serum. J Gene Med. 2004;6(6):681–692. | ||
Ueno S, Mojic M, Ohashi Y, Higashi N, Hayakawa Y, Irimura T. Asialoglycoprotein receptor promotes cancer metastasis by activating the EGFR–ERK pathway. Cancer Res. 2011;71(20):6419–6427. | ||
Khorev O, Stokmaier D, Schwardt O, Cutting B, Ernst B. Trivalent, Gal/GalNAc-containing ligands designed for the asialoglycoprotein receptor. Bioorg Med Chem. 2008;16(9):5216–5231. | ||
Geffen I, Spiess M. Asialoglycoprotein receptor. Int Rev Cytol. 1992; 137B:181–219. | ||
Sørensen AL, Clausen H, Wandall HH. Carbohydrate clearance receptors in transfusion medicine. Biochim Biophys Acta. 2012;1820(11):1797–1808. | ||
Avila MA, Berasain C, Sangro B, Prieto J. New therapies for hepatocellular carcinoma. Oncogene. 2006;25(27):3866–3884. | ||
Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29(36):4989–5005. | ||
Wang T, Upponi JR, Torchilin VP. Design of multifunctional non-viral gene vectors to overcome physiological barriers: dilemmas and strategies. Int J Pharm. 2012;427(1):3–20. | ||
Resina S, Prevot P, Thierry AR. Physico-chemical characteristics of lipoplexes influence cell uptake mechanisms and transfection efficacy. PLoS One. 2009;4(6):e6058. | ||
Harris RL, Van den Berg CW, Bowen DJ. ASGR1 and ASGR2, the genes that encode the asialoglycoprotein receptor (Ashwell Receptor), are expressed in peripheral blood monocytes and show interindividual differences in transcript profile. Mol Biol Int. 2012;2012:283–974. | ||
Hashida M, Nishikawa M, Yamashita F, Takakura Y. Cell-specific delivery of genes with glycosylated carriers. Adv Drug Deliv Rev. 2001;52(3):187–196. | ||
Faneca H, Cabrita AS, Simões S, Pedroso de Lima MC. Evaluation of the antitumoral effect mediated by IL-12 and HSV-tk genes when delivered by a novel lipid-based system. Biochim Biophys Acta. 2007;1768(5):1093–1102. | ||
Ma B, Zhang S, Jiang H, Zhao B, Lv H. Lipoplex morphologies and their influences on transfection efficiency in gene delivery. J Control Release. 2007;123(3):184–194. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.